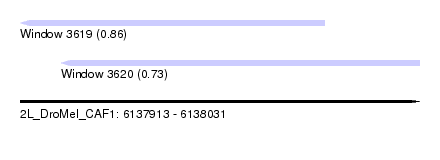
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,137,913 – 6,138,031 |
| Length | 118 |
| Max. P | 0.857954 |

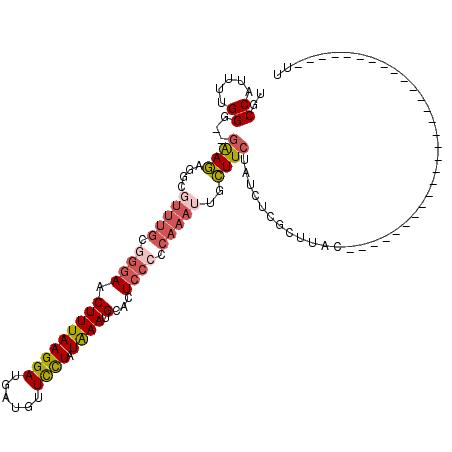
| Location | 6,137,913 – 6,138,003 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.42 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -9.18 |
| Energy contribution | -10.18 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6137913 90 - 22407834 UGCCAUUUUGGU--GAAGAGGCGUUUGCGGGAACUUUAAGGAUGAUGUUUCUAUAAAUGCACUCCCCCAAAUUGCUUCUAUCUCGAUUAC----------------------------UU .((...(((((.--(..((((((((((..(((((.(((....))).)))))..))))))).))).))))))..))...............----------------------------.. ( -22.40) >DroSec_CAF1 6454 90 - 1 UUCCAUUUUGGG--GAAGAGGCUUUUGCGGGAACUUUAAGGAUGAUGUUCCUAUAAAUGCACUCCACCAAAUUGCUUCUAUCUCGCUUAC----------------------------UU .........(((--(.((((((.((((..(((.(((((((((......)))).)))).)...)))..))))..)))))).))))......----------------------------.. ( -22.50) >DroSim_CAF1 6615 77 - 1 UGCCAUUUUGGG--GAAG-------------AACUUUAAGGAUGAUGUUCCUAUAAAUGCACUCUCCCAAAUUGCUUAUAUCUCGCUUAC----------------------------UU .((...((((((--..((-------------..(((((((((......)))).)))).)..))..))))))..))...............----------------------------.. ( -16.80) >DroEre_CAF1 6813 99 - 1 UGCCAUCUUGGGGGGGAAAGGUGUUUGUGGGAACUUUAAGGAUUCUGUUCCUAUAAAUGCACUCCCCCAAAU-GUUUCUUCCUAGCAUUC--------------------AAACAACUUU ........((((((((....((((((((((((((............)))))))))))))).))))))))(((-(((.......)))))).--------------------.......... ( -34.20) >DroYak_CAF1 6714 117 - 1 UGCCAUCCUGGG--GGAAAGACGUUUGUGGGAUCUUUAAGGAUGAUGUUCUUAUGAAUGCACUCCCCCAAAU-GUUUCUCUUUUGCAUGCCGAAAAUGAAUGUUAUCUCAAAACAACAUU ........((((--(((..(.((((..(((((...(((....)))...)))))..)))))..)))))))(((-(((((..(((((.....)))))..)))((((.......))))))))) ( -27.00) >consensus UGCCAUUUUGGG__GAAGAGGCGUUUGCGGGAACUUUAAGGAUGAUGUUCCUAUAAAUGCACUCCCCCAAAUUGCUUCUAUCUCGCUUAC____________________________UU ..((.....))...((((....(((((.((((.(((((((((......)))).)))).)...)))).)))))..)))).......................................... ( -9.18 = -10.18 + 1.00)

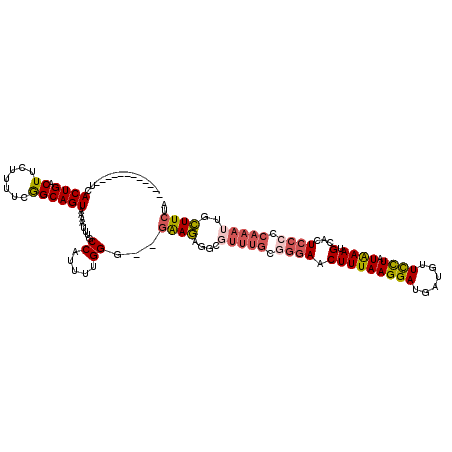
| Location | 6,137,925 – 6,138,031 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.74 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -12.38 |
| Energy contribution | -13.14 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6137925 106 - 22407834 ------------UCACUGACUUCUUUCGGGCAGUAAAAUUUGCCAUUUUGGU--GAAGAGGCGUUUGCGGGAACUUUAAGGAUGAUGUUUCUAUAAAUGCACUCCCCCAAAUUGCUUCUA ------------(((((((......)).(((((......))))).....)))--))(((((((((((.((((.(((((.(((.......))).)))).)...)))).)))).))))))). ( -29.10) >DroSec_CAF1 6466 103 - 1 ------------UCACUGACU---UUUCGGCAGUAAAAUUUUCCAUUUUGGG--GAAGAGGCUUUUGCGGGAACUUUAAGGAUGAUGUUCCUAUAAAUGCACUCCACCAAAUUGCUUCUA ------------.........---....((((((....((..((.....)).--.))(((((.((((..(((((.(((....))).)))))..)))).)).)))......)))))).... ( -25.60) >DroSim_CAF1 6627 93 - 1 ------------UCACUGACUUCUUUUCGGCAGUAAAAUUUGCCAUUUUGGG--GAAG-------------AACUUUAAGGAUGAUGUUCCUAUAAAUGCACUCUCCCAAAUUGCUUAUA ------------................(((((......)))))..((((((--..((-------------..(((((((((......)))).)))).)..))..))))))......... ( -21.90) >DroEre_CAF1 6833 119 - 1 GUGUCUUUGGUUUCACUGACUUAUUUUCAGCAGUAAAAUUUGCCAUCUUGGGGGGGAAAGGUGUUUGUGGGAACUUUAAGGAUUCUGUUCCUAUAAAUGCACUCCCCCAAAU-GUUUCUU ..(((..((....))..))).........((((......))))(((.(((((((((....((((((((((((((............)))))))))))))).)))))))))))-)...... ( -39.20) >DroYak_CAF1 6754 105 - 1 ------------UCACUGACUUCUUUUGAGCAGUAAAAUUUGCCAUCCUGGG--GGAAAGACGUUUGUGGGAUCUUUAAGGAUGAUGUUCUUAUGAAUGCACUCCCCCAAAU-GUUUCUC ------------..((((.(((.....))))))).........(((..((((--(((..(.((((..(((((...(((....)))...)))))..)))))..))))))).))-)...... ( -27.80) >consensus ____________UCACUGACUUCUUUUCGGCAGUAAAAUUUGCCAUUUUGGG__GAAGAGGCGUUUGCGGGAACUUUAAGGAUGAUGUUCCUAUAAAUGCACUCCCCCAAAUUGCUUCUA ..............((((.((.......))))))........((.....))...((((....(((((.((((.(((((((((......)))).)))).)...)))).)))))..)))).. (-12.38 = -13.14 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:16 2006