
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
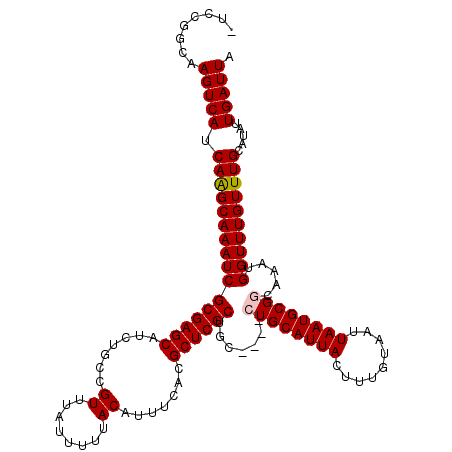
| Location | 6,136,995 – 6,137,113 |
| Length | 118 |
| Max. P | 0.985257 |

| Location | 6,136,995 – 6,137,113 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.36 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -27.80 |
| Energy contribution | -28.04 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
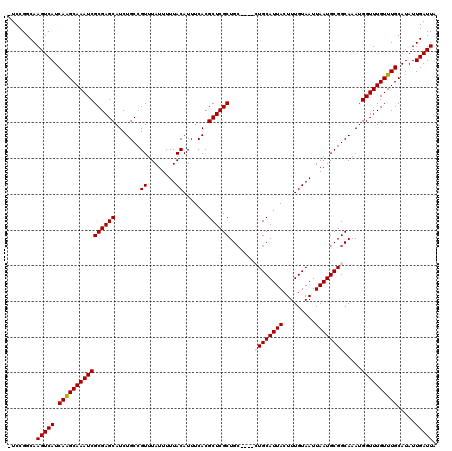
>2L_DroMel_CAF1 6136995 118 + 22407834 -UCCGGCAAGUCAUCAAGCAAAUCGCGAGCAUCUGCCGUUUAUUUUUACAUUUCACGCUCGCUGCUG-GCUGCAUUACUUUGUAAUUAAUGCGGCAAAUGGUUUGUUUGCAUAUUGAUUA -.......(((((.((((((((((((((((.......((........)).......)))))).....-(((((((((.........)))))))))....)))))))))).....))))). ( -33.94) >DroSec_CAF1 5502 115 + 1 CUCCGGCAAGUCAUCAAGCAAAUCGCGAGCAUCUGCCGUUUAUUUUUACAUUUCACGCUCGCUGC-----UGCAUUACUUUGUAAUUAAUGCGGCAAAUGGUUUGUUUGCAUAUUGAUUA ........(((((.((((((((((((((((.......((........)).......))))))(((-----(((((((.........))))))))))...)))))))))).....))))). ( -34.74) >DroSim_CAF1 5647 115 + 1 CUCCGGCAAGUCAUCAAGCAAAUCGCGAGCAUCUGCCGUUUAUUUUUACAUUUCACGCUCGCUGC-----UGCAUUACUUUGUAAUUAAUGCGGCAAAUGGUUUGUUUGCAUAUUGAUUA ........(((((.((((((((((((((((.......((........)).......))))))(((-----(((((((.........))))))))))...)))))))))).....))))). ( -34.74) >DroEre_CAF1 5886 114 + 1 -UCCGGCAAGUCAUCAAGCAAAUCGCGAGCAUCUGCCGUUUAUUUUUACAUUUCACGCUCGCUG-----CUGCAUUACUUUGUAAUUAAUGCGGCAAAUGGUUUGUUUGCAUAUUGAUUA -.......(((((.((((((((((((((((.......((........)).......))))))((-----((((((((.........))))))))))...)))))))))).....))))). ( -34.74) >DroYak_CAF1 5892 119 + 1 -UCCGGCAAGUCAUCAGGCAAAUCGCGAGCAUCUGCCGUUUAUUUUUACAUUUCACGCUCGCUGCCGCGCUGCAUUACUUUGUAAUUAAUGCGGCAAAUGGUUUGUUUGCAUAUUGAUUA -.......(((((.((((((((((((((((.......((........)).......))))))(((((((.((.(((((...))))))).)))))))...)))))))))).....))))). ( -35.34) >consensus _UCCGGCAAGUCAUCAAGCAAAUCGCGAGCAUCUGCCGUUUAUUUUUACAUUUCACGCUCGCUGC____CUGCAUUACUUUGUAAUUAAUGCGGCAAAUGGUUUGUUUGCAUAUUGAUUA ........(((((.((((((((((((((((.......((........)).......)))))).......((((((((.........)))))))).....)))))))))).....))))). (-27.80 = -28.04 + 0.24)

| Location | 6,136,995 – 6,137,113 |
|---|---|
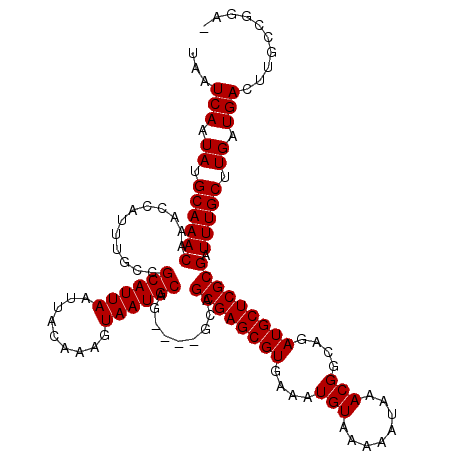
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.36 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
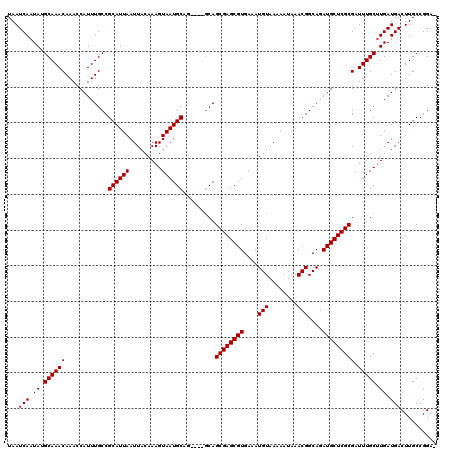
>2L_DroMel_CAF1 6136995 118 - 22407834 UAAUCAAUAUGCAAACAAACCAUUUGCCGCAUUAAUUACAAAGUAAUGCAGC-CAGCAGCGAGCGUGAAAUGUAAAAAUAAACGGCAGAUGCUCGCGAUUUGCUUGAUGACUUGCCGGA- ..(((((...((((((.........((.((((((.........)))))).))-.....((((((((....(((........)))....))))))))).))))))))))...........- ( -27.20) >DroSec_CAF1 5502 115 - 1 UAAUCAAUAUGCAAACAAACCAUUUGCCGCAUUAAUUACAAAGUAAUGCA-----GCAGCGAGCGUGAAAUGUAAAAAUAAACGGCAGAUGCUCGCGAUUUGCUUGAUGACUUGCCGGAG ..(((((...((((((........(((.((((((.........)))))).-----)))((((((((....(((........)))....))))))))).))))))))))............ ( -29.00) >DroSim_CAF1 5647 115 - 1 UAAUCAAUAUGCAAACAAACCAUUUGCCGCAUUAAUUACAAAGUAAUGCA-----GCAGCGAGCGUGAAAUGUAAAAAUAAACGGCAGAUGCUCGCGAUUUGCUUGAUGACUUGCCGGAG ..(((((...((((((........(((.((((((.........)))))).-----)))((((((((....(((........)))....))))))))).))))))))))............ ( -29.00) >DroEre_CAF1 5886 114 - 1 UAAUCAAUAUGCAAACAAACCAUUUGCCGCAUUAAUUACAAAGUAAUGCAG-----CAGCGAGCGUGAAAUGUAAAAAUAAACGGCAGAUGCUCGCGAUUUGCUUGAUGACUUGCCGGA- ..(((((...((((((........(((.((((((.........)))))).)-----))((((((((....(((........)))....))))))))).))))))))))...........- ( -29.00) >DroYak_CAF1 5892 119 - 1 UAAUCAAUAUGCAAACAAACCAUUUGCCGCAUUAAUUACAAAGUAAUGCAGCGCGGCAGCGAGCGUGAAAUGUAAAAAUAAACGGCAGAUGCUCGCGAUUUGCCUGAUGACUUGCCGGA- ...(((.((.((((((........((((((....(((((...))))).....))))))((((((((....(((........)))....))))))))).))))).)).))).........- ( -32.80) >consensus UAAUCAAUAUGCAAACAAACCAUUUGCCGCAUUAAUUACAAAGUAAUGCAG____GCAGCGAGCGUGAAAUGUAAAAAUAAACGGCAGAUGCUCGCGAUUUGCUUGAUGACUUGCCGGA_ ...(((.((.((((((............((((((.........)))))).........((((((((....(((........)))....))))))))).))))).)).))).......... (-25.10 = -25.10 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:14 2006