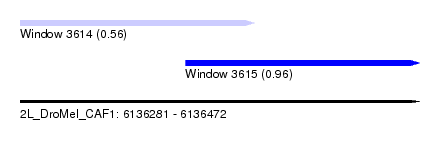
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,136,281 – 6,136,472 |
| Length | 191 |
| Max. P | 0.962641 |

| Location | 6,136,281 – 6,136,393 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 91.54 |
| Mean single sequence MFE | -29.31 |
| Consensus MFE | -26.32 |
| Energy contribution | -26.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
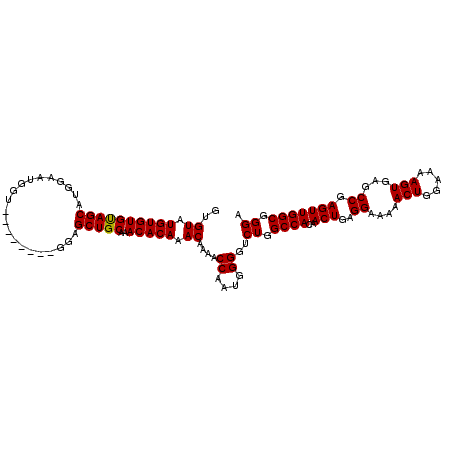
>2L_DroMel_CAF1 6136281 112 + 22407834 GUGUAUGUGUGUAGCAUGGAAUGGUGGCGCUGUGGAGCUGCAAACACAAACAAAACCAAUGGGG-CUGGCCACAACUGAGGAAAAACUGGAAAAGUGAGCCGAGUUGGCGGGA .(((.((((((((((.....((((.....))))...)))))..))))).)))...((....)).-((.((((..(((..((....(((.....)))...)).))))))).)). ( -29.50) >DroSec_CAF1 4898 105 + 1 GCGUAUGUGUGCAGCAUGGAAUGGU--------GGAGCUGCAAACACAAACAAAACCAAUGGGGUCUGGCCACAACUGAGGAAAAACUGGAAAAGUGAUCCGAGUUGGCGGGA ..((.((((((((((..........--------...)))))..))))).))....((....)).(((.((((..(((..(((...(((.....)))..))).))))))).))) ( -30.52) >DroSim_CAF1 4959 105 + 1 GUGUAUGUGUGUAGCAUGAAAUGGU--------GGAGCUGCAAACACAAACAAAACCAAUGGGGUCUGGCCACAACUGAGGAAAAACUGGAAAAGUGAGCCAAGUUGGCGGGA .(((.((((((((((..........--------...)))))..))))).)))...((....)).(((.((((..(((..((....(((.....)))...)).))))))).))) ( -27.92) >consensus GUGUAUGUGUGUAGCAUGGAAUGGU________GGAGCUGCAAACACAAACAAAACCAAUGGGGUCUGGCCACAACUGAGGAAAAACUGGAAAAGUGAGCCGAGUUGGCGGGA ..((.((((((((((.....................)))))..))))).))....((....))..((.((((..(((..((....(((.....)))...)).))))))).)). (-26.32 = -26.10 + -0.22)

| Location | 6,136,360 – 6,136,472 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -22.76 |
| Energy contribution | -22.60 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
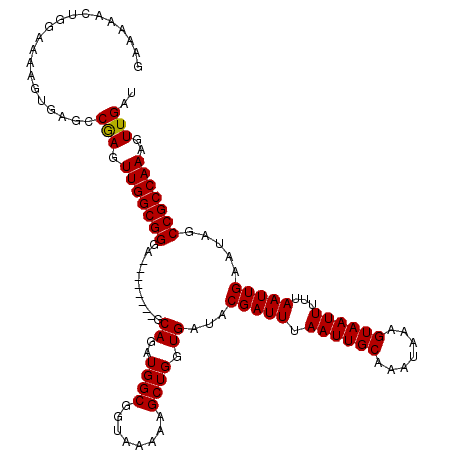
>2L_DroMel_CAF1 6136360 112 + 22407834 GAAAAACUGGAAAAGUGAGCCGAGUUGGCGGGA--------GCAGAUGGCGGUAAAAAGCUGGUGAUACGAUUUAAUUGCAAAUAAAGUAAUUUUUAAUUGAAUAGCCGCCAAAGUUGAU ....((((......((..(((.....)))....--------))...(((((((.......((......))(((((((((.((((......)))).))))))))).))))))).))))... ( -24.10) >DroSec_CAF1 4970 112 + 1 GAAAAACUGGAAAAGUGAUCCGAGUUGGCGGGA--------GCAGAUGGCGGUAAAAAGCUGGUGAUACGAUUUAAUUGCAAAUAAAGUAAUUUUUAAUUGAAUAGCCGCCAAAGUUGAU ....(((((((.......))).)))).((....--------))...(((((((.......((......))(((((((((.((((......)))).))))))))).)))))))........ ( -24.70) >DroSim_CAF1 5031 112 + 1 GAAAAACUGGAAAAGUGAGCCAAGUUGGCGGGA--------GCAGAUGGCGGUAAAAAGCUGGUGAUACGAUUUAAUUGCAAAUAAAGUAAUUUUUAAUUGAAUAGCCGCCAAAGUUGAU ....((((......((..(((.....)))....--------))...(((((((.......((......))(((((((((.((((......)))).))))))))).))))))).))))... ( -24.80) >DroEre_CAF1 5254 112 + 1 GAAAAACUGGAAAAUCGAGUCGAGUUGGCGGGA--------GCAGAUGGCGGUAAAAAGCUGGUGAUACGAUUUAAUUGCAAAUAAAGUAAUUUUUAAUUGAAUAGCCGCCAAAGUUGAU ..................(((((.(((((((..--------.((..((((........)))).))...(((((.((((((.......))))))...))))).....)))))))..))))) ( -24.80) >DroYak_CAF1 5266 120 + 1 GAAAAACUGGAAAAUCGAGCCGAGUUGGCGGGAGUUGGAGCGCAGAUGGCGGUAAAAAGCUGGUGAUACGAUUUAAUUGCAAAUAAAGUAAUUUUUAAUUGAAUGUCCGCCAAAGUUGAU ......((.(.....).)).(((.((((((((((((..(.(((.....))).)....)))).......(((((.((((((.......))))))...)))))....))))))))..))).. ( -26.90) >consensus GAAAAACUGGAAAAGUGAGCCGAGUUGGCGGGA________GCAGAUGGCGGUAAAAAGCUGGUGAUACGAUUUAAUUGCAAAUAAAGUAAUUUUUAAUUGAAUAGCCGCCAAAGUUGAU ....................(((.(((((((...........((..((((........)))).))...(((((.((((((.......))))))...))))).....)))))))..))).. (-22.76 = -22.60 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:11 2006