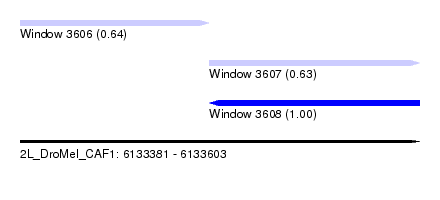
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,133,381 – 6,133,603 |
| Length | 222 |
| Max. P | 0.998879 |

| Location | 6,133,381 – 6,133,486 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 94.85 |
| Mean single sequence MFE | -20.39 |
| Consensus MFE | -19.74 |
| Energy contribution | -19.74 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6133381 105 + 22407834 UAAGCCUGCAAAGGAGCGAUUAAGAGUUUCGUUACAAAAAACACUCGUCCACUCGUCUUCUUCCUGUUUGUCUCGUCUCGUCUUUCGCUCCUACUAUCUUUCUCG ...........((((((((....(((...((..(((((.......((......))...........)))))..)).))).....))))))))............. ( -22.17) >DroSec_CAF1 2067 100 + 1 UAAGCCUGCAAAGGAGCGAUUAAGAGUUUCGUUACAAAAAACACUCGUCCACU---CUUCUUCCUGUUUGU--CGUCUCGCCCUUCGCUCCUACUAUCUUUCUCG ...........((((((((....(((...((..(((((........(......---).........)))))--)).))).....))))))))............. ( -18.13) >DroSim_CAF1 2065 102 + 1 UAAGCCUGCAAAGGAGCGAUUAAGAGUUUCGUUACAAAAAACACUCGUCCACU---CUUCUUCCUGUUUGUCUCGUCUCGUCCUUCGCUCCUACUAUCUUUCUCG ...........((((((((....(((...((..(((((........(......---).........)))))..)).))).....))))))))............. ( -20.63) >DroEre_CAF1 2141 102 + 1 UAAGCCUGCAAAGGAGCGAUUAAGAGUUUCGUUACAAAAAACACUCGUCGACU---CUUCAUCCUGUUUGUCUCGUCUCGCCUUUCGCUCCUACCAUCUUUCUCG ...........((((((((....(((...((..(((((........(......---).........)))))..)).))).....))))))))............. ( -20.63) >consensus UAAGCCUGCAAAGGAGCGAUUAAGAGUUUCGUUACAAAAAACACUCGUCCACU___CUUCUUCCUGUUUGUCUCGUCUCGCCCUUCGCUCCUACUAUCUUUCUCG ...........((((((((....(((...((..(((((............................)))))..)).))).....))))))))............. (-19.74 = -19.74 + 0.00)

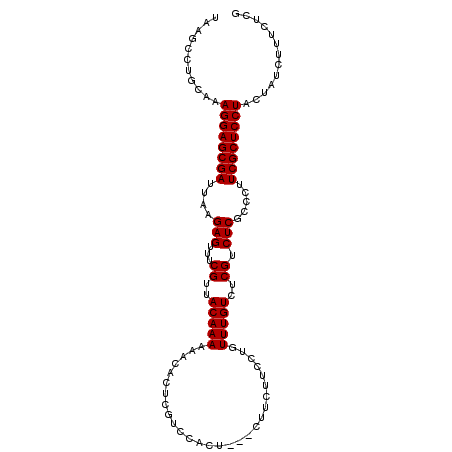
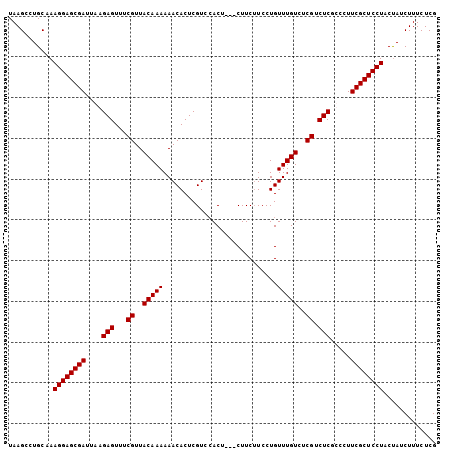
| Location | 6,133,486 – 6,133,603 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.26 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -24.48 |
| Energy contribution | -24.52 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6133486 117 + 22407834 UUACAUUUAUUGGCGUGUUUGUGCAUAAUGAAGCAAAGUCCAUUUUUGUGGUUAGCUUCGGAUGCUCUCUCAUUUCUGUU---GCUCCCAACAUGCAAAUGAAAUGGCCAUUCAUUUGCG ..........((((......(.((((..((((((.....((((....))))...)))))).)))).)...((((((..((---((.........))))..)))))))))).......... ( -29.70) >DroSec_CAF1 2167 117 + 1 UUACAUUUAUUGGCGUGUUUGUGCAUAAUGAAGCAAAGUCCAUUUUUGUGGUUAGCUUCUGAUGUUCUCUCAUUUCUGUU---GGCCCCAACAUGCAAAUGAAAUGGUCAUUCAUUUGCG ...........((.(((...(.((((...(((((.....((((....))))...)))))..)))).)...))).))((((---(....))))).(((((((((.......))))))))). ( -30.20) >DroSim_CAF1 2167 117 + 1 UUACAUUUAUUGGCGUGUUUGUGCAUAAUGAAGCAAAGUCCAUUUUUGUGGUUAGCUUCUGAUGUUCUCUCAUUUCUGUU---GCCCCCAACAUGCAAAUGAAAUGGCCAUUCAUUUGCG ...........((.(((...(.((((...(((((.....((((....))))...)))))..)))).)...))).))((((---(....))))).(((((((((.......))))))))). ( -28.00) >DroEre_CAF1 2243 117 + 1 UUACAUUUAUUGGCGUGUUUGUGCAUAAUGAAGCAAAGUCCAUUUUUGUGGUUAGCUUCGGAUAUUCUCUCAUUUCUGUU---GGCCCCAACAUGCAAAUGAAAUGGCCAUUCAUUUGCG ..........((((((((....))))..((((((.....((((....))))...)))))).........(((((((((((---(....))))).).))))))....)))).......... ( -27.00) >DroYak_CAF1 2305 120 + 1 UUACAUUUAUUGGCGUGUUUGUGCAUAAUGAAGCAAAGUCCAUUUUUGUGGUUAGCUUCGGAUGUUCUCUCAUUUCUGUUUUUACCCUUAGCAUGCAAAUGAAAUGGCCAUUCAUUUGCG ....................((((.....(((((.....((((....))))...)))))(((((......)))))...............))))(((((((((.......))))))))). ( -25.40) >consensus UUACAUUUAUUGGCGUGUUUGUGCAUAAUGAAGCAAAGUCCAUUUUUGUGGUUAGCUUCGGAUGUUCUCUCAUUUCUGUU___GCCCCCAACAUGCAAAUGAAAUGGCCAUUCAUUUGCG ...........((.(((...(.((((...(((((.....((((....))))...)))))..)))).)...))).))..................(((((((((.......))))))))). (-24.48 = -24.52 + 0.04)

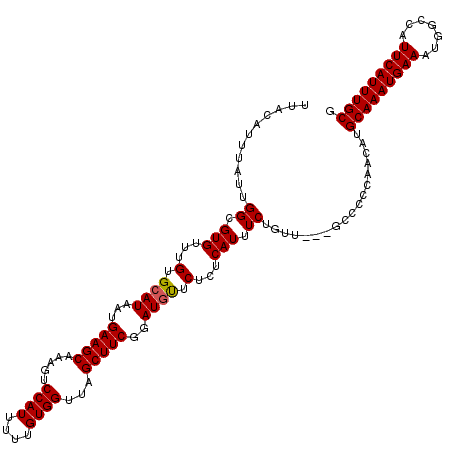
| Location | 6,133,486 – 6,133,603 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.26 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -24.38 |
| Energy contribution | -25.18 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
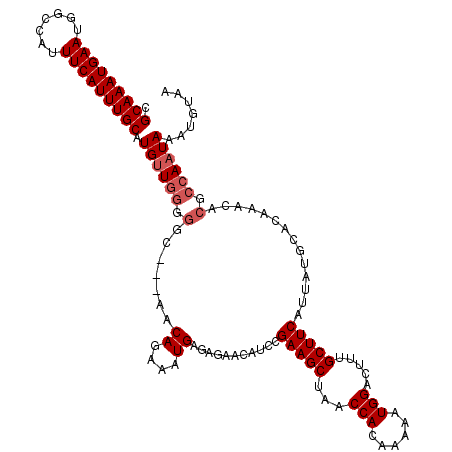
>2L_DroMel_CAF1 6133486 117 - 22407834 CGCAAAUGAAUGGCCAUUUCAUUUGCAUGUUGGGAGC---AACAGAAAUGAGAGAGCAUCCGAAGCUAACCACAAAAAUGGACUUUGCUUCAUUAUGCACAAACACGCCAAUAAAUGUAA .(((((((((.......))))))))).((((((....---..((....)).....((((..(((((...(((......))).....)))))...)))).........))))))....... ( -30.20) >DroSec_CAF1 2167 117 - 1 CGCAAAUGAAUGACCAUUUCAUUUGCAUGUUGGGGCC---AACAGAAAUGAGAGAACAUCAGAAGCUAACCACAAAAAUGGACUUUGCUUCAUUAUGCACAAACACGCCAAUAAAUGUAA .(((((((((.......))))))))).((((((.(..---..((....))...........(((((...(((......))).....)))))..............).))))))....... ( -27.70) >DroSim_CAF1 2167 117 - 1 CGCAAAUGAAUGGCCAUUUCAUUUGCAUGUUGGGGGC---AACAGAAAUGAGAGAACAUCAGAAGCUAACCACAAAAAUGGACUUUGCUUCAUUAUGCACAAACACGCCAAUAAAUGUAA .(((((((((.......))))))))).((((((.(..---..((....))...........(((((...(((......))).....)))))..............).))))))....... ( -28.60) >DroEre_CAF1 2243 117 - 1 CGCAAAUGAAUGGCCAUUUCAUUUGCAUGUUGGGGCC---AACAGAAAUGAGAGAAUAUCCGAAGCUAACCACAAAAAUGGACUUUGCUUCAUUAUGCACAAACACGCCAAUAAAUGUAA .(((((((((.......))))))))).((((((.(..---..((....))...........(((((...(((......))).....)))))..............).))))))....... ( -27.80) >DroYak_CAF1 2305 120 - 1 CGCAAAUGAAUGGCCAUUUCAUUUGCAUGCUAAGGGUAAAAACAGAAAUGAGAGAACAUCCGAAGCUAACCACAAAAAUGGACUUUGCUUCAUUAUGCACAAACACGCCAAUAAAUGUAA .(((((((((.......))))))))).(((...((((.....((....)).......))))(((((...(((......))).....))))).....)))..................... ( -25.00) >consensus CGCAAAUGAAUGGCCAUUUCAUUUGCAUGUUGGGGGC___AACAGAAAUGAGAGAACAUCCGAAGCUAACCACAAAAAUGGACUUUGCUUCAUUAUGCACAAACACGCCAAUAAAUGUAA .(((((((((.......))))))))).((((((.(.......((....))...........(((((...(((......))).....)))))..............).))))))....... (-24.38 = -25.18 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:04 2006