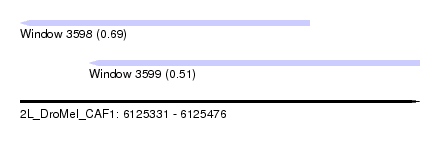
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,125,331 – 6,125,476 |
| Length | 145 |
| Max. P | 0.685518 |

| Location | 6,125,331 – 6,125,436 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.12 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -25.69 |
| Energy contribution | -25.50 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
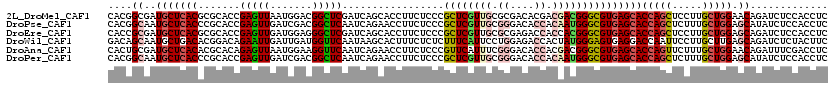
>2L_DroMel_CAF1 6125331 105 - 22407834 AUCAGCACCUUCUCCCGCUCGUUGCGCGACACGACGACGGGCGUGAGCACCAGCUCCUUGCUGGAACAGAUCUCCACCUCCGGCUUGUUGUGCCGCUCCACCACC--------------- ...............(((((((((((.....)).))))))))).((((.(((((.....))))).................(((.......))))))).......--------------- ( -34.90) >DroPse_CAF1 108567 105 - 1 AUCAGAACCUUCUCCCGCUCGUUGCGGGACACCACAAUGGGCGUGAGCACCAGCUCUUUGCUGGAGCAUAUCUCCACCUCCGGCUUGUUGUGACGCUCCACCACU--------------- .(((.(((.......(((((((((.((....)).))))))))).((((....))))...(((((((...........)))))))..))).)))............--------------- ( -35.70) >DroSim_CAF1 43492 105 - 1 AUCAGCACCUUCUCCCGCUCGUUGCGCGACACCACGACGGGCGUGAGCACCAGCUCCUUGCUGGAGCAGAUCUCCACCUCCGGUUUGUUGUGCCGCUCCACAACC--------------- ....((((...(((.(((((((((.(......).))))))))).)))..(((((.....)))))((((((((.........))))))))))))............--------------- ( -35.90) >DroWil_CAF1 113586 120 - 1 AUAAGCACUUUCUCUCUUUCAUUCCUGGAGACCACUAUGGGAGUGAGGACCAAUUCCUUGCUUGAGCAGAUCUCUACUUCGGGUUUGUUGUGUCGUUCCACUACCUAAGAAAUGGAAAUG ........(((((.((((.......(((((..(((.....(((..((((.....))))..))).(((((((((.......))))))))))))...)))))......))))...))))).. ( -28.42) >DroAna_CAF1 31587 105 - 1 AUCAGAACCUUCUCCCGUUCAUUUCGGGACACCACGACGGGCGUGAGCACCAGUUCUUUGCUGGAACAGAUUUCGACCUCCGGCUUGUUGUGGCGCUCCACGACC--------------- ....((((........))))...(((((((.(((((((((((....)).))........((((((...(....)....))))))..))))))).).))).)))..--------------- ( -37.10) >DroPer_CAF1 106730 105 - 1 AUCAGAACCUUCUCCCGCUCGUUGCGGGACACCACAAUGGGCGUGAGCACCAGCUCUUUGCUGGAGCAUAUCUCCACCUCCGGCUUGUUGUGACGCUCCACCACU--------------- .(((.(((.......(((((((((.((....)).))))))))).((((....))))...(((((((...........)))))))..))).)))............--------------- ( -35.70) >consensus AUCAGAACCUUCUCCCGCUCGUUGCGGGACACCACGACGGGCGUGAGCACCAGCUCCUUGCUGGAGCAGAUCUCCACCUCCGGCUUGUUGUGACGCUCCACCACC_______________ ...............(((((((((.((....)).))))))))).((((.(((((.....(((((((...........)))))))..)))).)..))))...................... (-25.69 = -25.50 + -0.19)

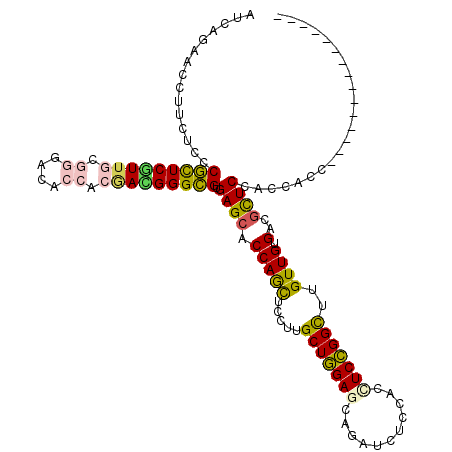
| Location | 6,125,356 – 6,125,476 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.33 |
| Mean single sequence MFE | -43.07 |
| Consensus MFE | -29.13 |
| Energy contribution | -30.17 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
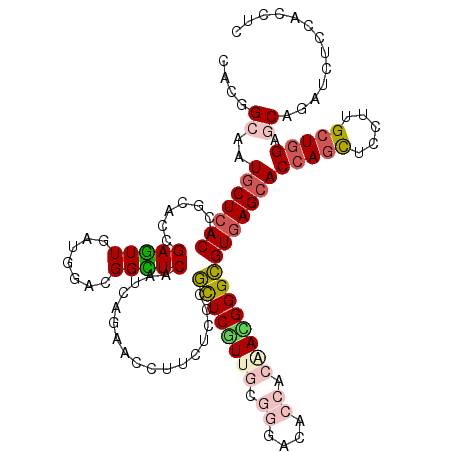
>2L_DroMel_CAF1 6125356 120 - 22407834 CACGGCGAUGCUCACGCGCACCGAGUUAAUGGACGGCUCGAUCAGCACCUUCUCCCGCUCGUUGCGCGACACGACGACGGGCGUGAGCACCAGCUCCUUGCUGGAACAGAUCUCCACCUC ...((.(((((((((((((..((((((.......))))))....))........(((.((((((.......))))))))))))))))).(((((.....))))).....))).))..... ( -47.80) >DroPse_CAF1 108592 120 - 1 CACGGCAAUGCUCACCCGCACCGAGUUGAUCGACGGCUCAAUCAGAACCUUCUCCCGCUCGUUGCGGGACACCACAAUGGGCGUGAGCACCAGCUCUUUGCUGGAGCAUAUCUCCACCUC ....((..(((((((..((.((((((((.....)))))).............((((((.....)))))).........)))))))))))(((((.....))))).))............. ( -44.10) >DroEre_CAF1 52454 120 - 1 CACCGCGAUGCUCACGCGCACCGAGUUGAUGGAGGGCUCGAUCAGCACCUUCUCCCGCUCGUUGCGCGAGACCACCACGGGCGUGAGCACCAGCUCCUUGCUGGAGCAGAUCUCCACCUC ....((..((((((((((((.(((((.(..((((((((.....))).)))))..).))))).)))))....((.....))..)))))))(((((.....))))).))............. ( -52.50) >DroWil_CAF1 113626 120 - 1 GACAGCAAUGCUGACACGGACAGAAUUGAUUGAUGGUUCAAUAAGCACUUUCUCUCUUUCAUUCCUGGAGACCACUAUGGGAGUGAGGACCAAUUCCUUGCUUGAGCAGAUCUCUACUUC ..((((...))))....(((..(((((((((((....))))).......(((((.((..(((....(....)....)))..)).))))).)))))).((((....))))...)))..... ( -28.50) >DroAna_CAF1 31612 120 - 1 CACUGCGAUGCUCACACGCACAGAGUUAAUGGAAGGUUCAAUCAGAACCUUCUCCCGUUCAUUUCGGGACACCACGACGGGCGUGAGCACCAGUUCUUUGCUGGAACAGAUUUCGACCUC ..(((...(((((((.(.(............((((((((.....))))))))(((((.......))))).........).).)))))))(((((.....)))))..)))........... ( -41.40) >DroPer_CAF1 106755 120 - 1 CACGGCAAUGCUCACCCGCACCGAGUUGAUCGACGGCUCAAUCAGAACCUUCUCCCGCUCGUUGCGGGACACCACAAUGGGCGUGAGCACCAGCUCUUUGCUGGAGCAUAUCUCCACCUC ....((..(((((((..((.((((((((.....)))))).............((((((.....)))))).........)))))))))))(((((.....))))).))............. ( -44.10) >consensus CACGGCAAUGCUCACACGCACCGAGUUGAUGGACGGCUCAAUCAGAACCUUCUCCCGCUCGUUGCGGGACACCACAACGGGCGUGAGCACCAGCUCCUUGCUGGAGCAGAUCUCCACCUC ....((..(((((((.......(((((.......))))).................((((((((.((....)).)))))))))))))))(((((.....))))).))............. (-29.13 = -30.17 + 1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:55 2006