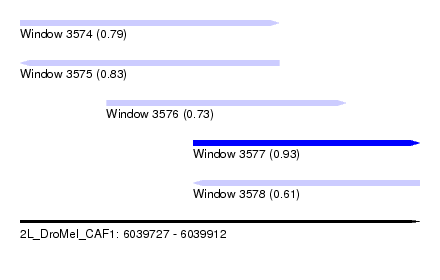
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,039,727 – 6,039,912 |
| Length | 185 |
| Max. P | 0.927872 |

| Location | 6,039,727 – 6,039,847 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -40.95 |
| Consensus MFE | -19.02 |
| Energy contribution | -19.72 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6039727 120 + 22407834 UUGGCAUUCAGGAUUGCUUUCUCCUCAGUCGCGGCAUGCAAAUAGCCAGCGAACUAAGGUUUCGCCAGCAUCUUCCGGCAUUAGCUGGCAUGCAACUGUCCACAAUGGACAGCAUUCUCA .((((....((((.....))))(((.(((((((((((....)).))).))).))).)))....))))((((...(((((....))))).))))..((((((.....))))))........ ( -41.00) >DroVir_CAF1 3314 120 + 1 UUGAUAUACAGGAUUGUUUUCUGCUCGGCCGCGGCAUGCAGAUCGCCUGCGAGCUGCGCUUCCGCCAACACGUGCCGCAUCUGCUGGCCAUGCAGCUGUCCACCAUGGACAGCAUUCUGA ........((((((.......(((..(((((((((((((((.(((....))).))))((....))......))))))).......))))..)))(((((((.....))))))))))))). ( -51.11) >DroGri_CAF1 3611 120 + 1 UUGACAUCCAGGAUUGCUUCCAGCUGAGUCGCGGCAUGCAGAUCGCCAGCGAGCUGCGCUUUCGCCAGCAUGUGCCGCAUCUGCUGGCCAUGCAACUGUCCACAAUGGACAGCAUACUGA ........(((..((((..(((((.((...(((((((((((.(((....))).))))(((......)))..))))))).)).)))))....))))((((((.....))))))....))). ( -50.80) >DroWil_CAF1 16 120 + 1 UUGAUAUACAGGAUUGCUUUCUGCUUAGUCGUGGCAUGCAAAUAGCCUGUGAAUUGCGUUUCCGUCACAAUGUGCCUCCAUUAAUGACCAUGCAGUUGACCACAAUGGAUAGUAUUCUAA (((...((((((.((((....((((.......)))).))))....))))))((((((((....((((.((((......))))..)))).)))))))).....)))((((......)))). ( -26.30) >DroYak_CAF1 2631 120 + 1 UUGCUAUCCAGGAUUGCUUUCUACUCAGUCGGGGCAUGCAAAUAGCCAGCGAACUACGCUUUCGCCAGCAUCUUCCGUCACUAAUUGGCAUGCAGCUCUCCACUAUGGACAGCAUUCUCA .((((.(((((((..(((.....(((....)))((((((.....((..(((((.......)))))..))......((........)))))))))))..)))....)))).))))...... ( -33.80) >DroMoj_CAF1 3484 120 + 1 UCAAUAUCCAGGAUUGUUUCCUACUCAGCCGUGGCAUGCAGAUUGCUUGCGAGCUGCGCUUCCGUCAGCAUGUACCACAUGUGCUGGCCAUGCAGCUGGCCACCAUGGACAGCAUCCUAA .........(((((.((((((......(((...(((.((.....)).))).(((((((.....((((((((((.....))))))))))..))))))))))......))).)))))))).. ( -42.70) >consensus UUGAUAUCCAGGAUUGCUUUCUGCUCAGUCGCGGCAUGCAAAUAGCCAGCGAACUGCGCUUCCGCCAGCAUGUGCCGCAACUACUGGCCAUGCAGCUGUCCACAAUGGACAGCAUUCUAA .........((((.....))))....(((((((((.........))).))).)))............(((((.((((.......))))))))).(((((((.....)))))))....... (-19.02 = -19.72 + 0.70)

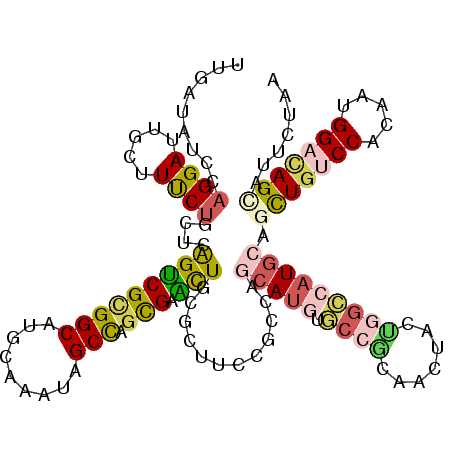
| Location | 6,039,727 – 6,039,847 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -44.89 |
| Consensus MFE | -18.50 |
| Energy contribution | -20.08 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6039727 120 - 22407834 UGAGAAUGCUGUCCAUUGUGGACAGUUGCAUGCCAGCUAAUGCCGGAAGAUGCUGGCGAAACCUUAGUUCGCUGGCUAUUUGCAUGCCGCGACUGAGGAGAAAGCAAUCCUGAAUGCCAA .......(((((((.....)))))))(((.(((((((......(....)..)))))))...(((((((.(((.(((.........))))))))))))).....))).............. ( -45.10) >DroVir_CAF1 3314 120 - 1 UCAGAAUGCUGUCCAUGGUGGACAGCUGCAUGGCCAGCAGAUGCGGCACGUGUUGGCGGAAGCGCAGCUCGCAGGCGAUCUGCAUGCCGCGGCCGAGCAGAAAACAAUCCUGUAUAUCAA .(((...(((((((.....)))))))(((.(((((.......((((((.(.((((((....)).)))).)((((.....)))).))))))))))).)))..........)))........ ( -51.71) >DroGri_CAF1 3611 120 - 1 UCAGUAUGCUGUCCAUUGUGGACAGUUGCAUGGCCAGCAGAUGCGGCACAUGCUGGCGAAAGCGCAGCUCGCUGGCGAUCUGCAUGCCGCGACUCAGCUGGAAGCAAUCCUGGAUGUCAA ((((...(((((((.....)))))))(((....(((((((.(((((((.......((....))((((.(((....))).)))).))))))).))..)))))..)))...))))....... ( -52.10) >DroWil_CAF1 16 120 - 1 UUAGAAUACUAUCCAUUGUGGUCAACUGCAUGGUCAUUAAUGGAGGCACAUUGUGACGGAAACGCAAUUCACAGGCUAUUUGCAUGCCACGACUAAGCAGAAAGCAAUCCUGUAUAUCAA ((((.......(((((((((..((......))..)).)))))))((((..((((((((....))....))))))((.....)).))))....))))((((.........))))....... ( -32.10) >DroYak_CAF1 2631 120 - 1 UGAGAAUGCUGUCCAUAGUGGAGAGCUGCAUGCCAAUUAGUGACGGAAGAUGCUGGCGAAAGCGUAGUUCGCUGGCUAUUUGCAUGCCCCGACUGAGUAGAAAGCAAUCCUGGAUAGCAA ......(((((((((....((((((((((........(((((.(....).)))))((....)))))))))(((..((((((.............))))))..)))..)))))))))))). ( -43.82) >DroMoj_CAF1 3484 120 - 1 UUAGGAUGCUGUCCAUGGUGGCCAGCUGCAUGGCCAGCACAUGUGGUACAUGCUGACGGAAGCGCAGCUCGCAAGCAAUCUGCAUGCCACGGCUGAGUAGGAAACAAUCCUGGAUAUUGA (((((((....(((.((.((((((......)))))).))..(((((((...((((.((....))))))..(((.......))).)))))))........)))....)))))))....... ( -44.50) >consensus UCAGAAUGCUGUCCAUUGUGGACAGCUGCAUGGCCAGCAGAGGCGGCACAUGCUGGCGAAAGCGCAGCUCGCAGGCUAUCUGCAUGCCGCGACUGAGCAGAAAGCAAUCCUGGAUAUCAA ...(...(((((((.....)))))))..).............((((((...((((.((....))))))......((.....)).)))))).............................. (-18.50 = -20.08 + 1.59)



| Location | 6,039,767 – 6,039,878 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -23.57 |
| Energy contribution | -23.68 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6039767 111 + 22407834 AAUAGCCAGCGAACUAAGGUUUCGCCAGCAUCUUCCGGCAUUAGCUGGCAUGCAACUGUCCACAAUGGACAGCAUUCUCAUCGGUUGUGCGGAGAGGCACUUGAGAGAUGC ....((((((((((....)))).(((..........)))....))))))..(((.((((((.....))))))...((((.(((...((((......)))).)))))))))) ( -38.90) >DroVir_CAF1 3354 111 + 1 GAUCGCCUGCGAGCUGCGCUUCCGCCAACACGUGCCGCAUCUGCUGGCCAUGCAGCUGUCCACCAUGGACAGCAUUCUGAUUGCCUGUGCCGAACGGCAUUUGGGCAACAG ....(((.((((((.((((............)))).)).)).)).)))......(((((((.....)))))))...(((.(((((((((((....)))))..))))))))) ( -46.00) >DroPse_CAF1 1410 111 + 1 AAUAGCCUGCGAGCUAAGAUUUCGUCAGGAUACACCUGCUUUGGUGGCCAUGCAGCUAUCUACGAUGGAUAGCAUUCUCAUUGGAUGUGCGGAGCGGCAUUUGGCCAAUGC .....((((((((.......)))).))))...((((......))))((..(((((((((((.....)))))))((((.....)))).))))..))(((.....)))..... ( -35.80) >DroEre_CAF1 2676 111 + 1 AAUAGCCAGCGAACUACGAUUUCGCCAGCAUCUUCCCCAACUAGCUGGCAUGCAGCUGUCCACUAUGGACAGCAUUCUCAUCGGUUGUGCAGAGAGGCACUUGAGGGAUGC ....(((((((((.......)))(....)..............))))))..((((((((((.....)))))))..((((.(((...((((......)))).)))))))))) ( -36.40) >DroYak_CAF1 2671 111 + 1 AAUAGCCAGCGAACUACGCUUUCGCCAGCAUCUUCCGUCACUAAUUGGCAUGCAGCUCUCCACUAUGGACAGCAUUCUCAUUGCUUGUGCUGAGAGGCACUUGAGUGAUGC ....((..(((((.......)))))..))......(((((((.....((.....(((.(((.....))).))).((((((..((....)))))))))).....))))))). ( -33.70) >DroPer_CAF1 1410 111 + 1 AAUAGCCUGCGAGCUAAGAUUUCGUCAGGAUACACCUGCUUUGGUGGCCAUGCAGCUAUCUACGAUGGAUAGCAUUCUCAUUGGAUGUGCGGAGCGGCAUUUGGCCAAUGC .....((((((((.......)))).))))...((((......))))((..(((((((((((.....)))))))((((.....)))).))))..))(((.....)))..... ( -35.80) >consensus AAUAGCCAGCGAACUAAGAUUUCGCCAGCAUCUUCCGGCACUAGUGGCCAUGCAGCUGUCCACGAUGGACAGCAUUCUCAUUGGUUGUGCGGAGAGGCACUUGAGCAAUGC ....((..(((((.......)))))..)).........................(((((((.....))))))).....((((((((((((......))))..)))))))). (-23.57 = -23.68 + 0.11)

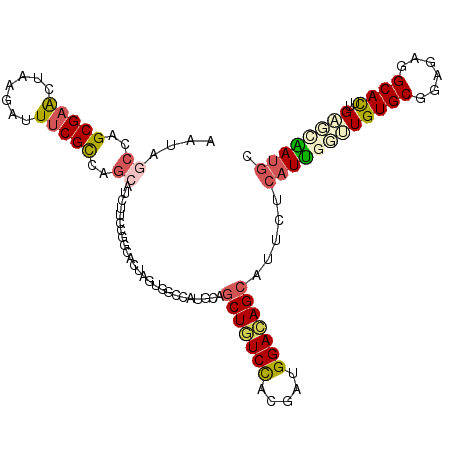
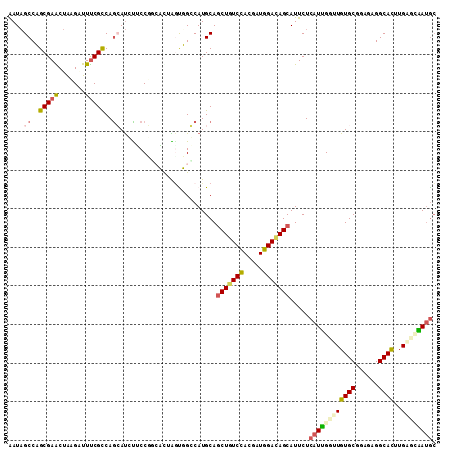
| Location | 6,039,807 – 6,039,912 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.51 |
| Mean single sequence MFE | -38.47 |
| Consensus MFE | -21.84 |
| Energy contribution | -21.15 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6039807 105 + 22407834 UUAGCUGGCAUGCAACUGUCCACAAUGGACAGCAUUCUCAUCGGUUGUGCGGAGAGGCACUUGAGAGAUGCCGAAAAGGACCCUCUAAAUGCCAGCGAAUUGAGC ...((((((((((..((((((.....))))))....(((..((......))..))))).....((((...((.....))...))))..))))))))......... ( -37.70) >DroVir_CAF1 3394 105 + 1 CUGCUGGCCAUGCAGCUGUCCACCAUGGACAGCAUUCUGAUUGCCUGUGCCGAACGGCAUUUGGGCAACAGCGAACAGCAACCGCUAAAUGCCAGCGAUCUGAGC .(((((((..(((.(((((((.....)))))))....((.(((((((((((....)))))..)))))))))))...(((....)))....)))))))........ ( -46.00) >DroPse_CAF1 1450 105 + 1 UUGGUGGCCAUGCAGCUAUCUACGAUGGAUAGCAUUCUCAUUGGAUGUGCGGAGCGGCAUUUGGCCAAUGCAGAACGAGAGCCACUAACUGCGAGUGAGCUCAGC (((((((((.....(((((((.....))))))).((((((((((..((((......))))....)))))).))))...).))))))))....(((....)))... ( -39.10) >DroYak_CAF1 2711 105 + 1 CUAAUUGGCAUGCAGCUCUCCACUAUGGACAGCAUUCUCAUUGCUUGUGCUGAGAGGCACUUGAGUGAUGCCGAAAAGGACCCUCUGAAUGCUAGCGAAUUGAGC .(((((.((.........(((.....))).(((((((.(((..((((((((....)))))..)))..)))((.....)).......))))))).)).)))))... ( -33.50) >DroMoj_CAF1 3564 105 + 1 GUGCUGGCCAUGCAGCUGGCCACCAUGGACAGCAUCCUAAUUGCCUGUGCCGAGCGCCACUUGAGCAAUAGCGAGGAGCAGCCCCUCAAUGCCAGCGAUCUGAGC .(((((((...((.((((.((.....)).))))......((((((.(((.(....).)))..).))))).))((((.......))))...)))))))........ ( -35.40) >DroPer_CAF1 1450 105 + 1 UUGGUGGCCAUGCAGCUAUCUACGAUGGAUAGCAUUCUCAUUGGAUGUGCGGAGCGGCAUUUGGCCAAUGCAGAACGAGAGCCACUAACUGCGAGUGAGCUCAGC (((((((((.....(((((((.....))))))).((((((((((..((((......))))....)))))).))))...).))))))))....(((....)))... ( -39.10) >consensus UUGGUGGCCAUGCAGCUGUCCACCAUGGACAGCAUUCUCAUUGCAUGUGCGGAGCGGCACUUGAGCAAUGCCGAACAGGAGCCACUAAAUGCCAGCGAACUGAGC .....(((......(((((((.....))))))).....((((((..((((......))))....))))))..........))).......(((((....))).)) (-21.84 = -21.15 + -0.69)

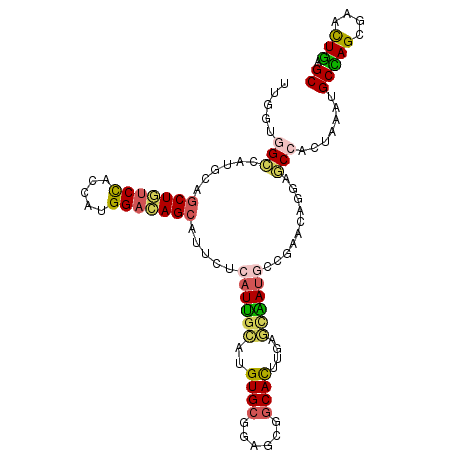
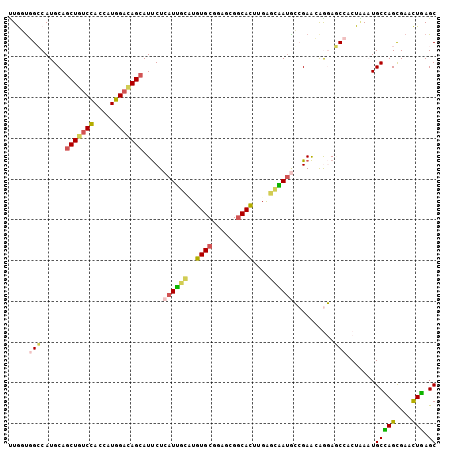
| Location | 6,039,807 – 6,039,912 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.51 |
| Mean single sequence MFE | -36.32 |
| Consensus MFE | -19.47 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6039807 105 - 22407834 GCUCAAUUCGCUGGCAUUUAGAGGGUCCUUUUCGGCAUCUCUCAAGUGCCUCUCCGCACAACCGAUGAGAAUGCUGUCCAUUGUGGACAGUUGCAUGCCAGCUAA .........((((((((...(((((.((.....))...)))))..((((......)))).........(...(((((((.....)))))))..)))))))))... ( -37.60) >DroVir_CAF1 3394 105 - 1 GCUCAGAUCGCUGGCAUUUAGCGGUUGCUGUUCGCUGUUGCCCAAAUGCCGUUCGGCACAGGCAAUCAGAAUGCUGUCCAUGGUGGACAGCUGCAUGGCCAGCAG .........((((((....(((((.......)))))((((((....((((....))))..))))))((....(((((((.....)))))))....)))))))).. ( -44.40) >DroPse_CAF1 1450 105 - 1 GCUGAGCUCACUCGCAGUUAGUGGCUCUCGUUCUGCAUUGGCCAAAUGCCGCUCCGCACAUCCAAUGAGAAUGCUAUCCAUCGUAGAUAGCUGCAUGGCCACCAA ((.(((....))))).....((((((...(((((.((((((.....(((......)))...)))))))))))((((((.......)))))).....))))))... ( -32.00) >DroYak_CAF1 2711 105 - 1 GCUCAAUUCGCUAGCAUUCAGAGGGUCCUUUUCGGCAUCACUCAAGUGCCUCUCAGCACAAGCAAUGAGAAUGCUGUCCAUAGUGGAGAGCUGCAUGCCAAUUAG ((((.......((((((((.((((((((.....)).))).)))..((((......)))).........))))))))(((.....))))))).............. ( -31.00) >DroMoj_CAF1 3564 105 - 1 GCUCAGAUCGCUGGCAUUGAGGGGCUGCUCCUCGCUAUUGCUCAAGUGGCGCUCGGCACAGGCAAUUAGGAUGCUGUCCAUGGUGGCCAGCUGCAUGGCCAGCAC ((.(((....)))))......((((.((((((....((((((...(((.(....).))).)))))).)))).)).)))).((.((((((......)))))).)). ( -40.90) >DroPer_CAF1 1450 105 - 1 GCUGAGCUCACUCGCAGUUAGUGGCUCUCGUUCUGCAUUGGCCAAAUGCCGCUCCGCACAUCCAAUGAGAAUGCUAUCCAUCGUAGAUAGCUGCAUGGCCACCAA ((.(((....))))).....((((((...(((((.((((((.....(((......)))...)))))))))))((((((.......)))))).....))))))... ( -32.00) >consensus GCUCAGAUCGCUGGCAUUUAGAGGCUCCUGUUCGGCAUUGCCCAAAUGCCGCUCCGCACAACCAAUGAGAAUGCUGUCCAUCGUGGACAGCUGCAUGGCCACCAA ((.(((....))))).......((((.........(((((((....(((......)))..))))))).(...(((((((.....)))))))..)..))))..... (-19.47 = -20.25 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:35 2006