| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,983,996 – 5,984,115 |
| Length | 119 |
| Max. P | 0.735236 |

| Location | 5,983,996 – 5,984,115 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.93 |
| Mean single sequence MFE | -36.66 |
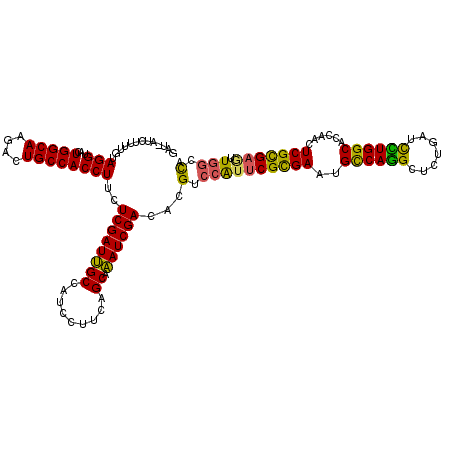
| Consensus MFE | -29.87 |
| Energy contribution | -28.85 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.735236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5983996 119 + 22407834 AAU-CUCCUCCGUAGGAACUGGCAAGACUGCCACCUUCUCGAUUGCUAUCCUUCAGCAAAUCGACACGAGCAUUCGCGAGUGCCAGGCGCUGAUCCUGGCCCCCACUCGCGAGUUGGCCA ...-......(((.(((..(((((....)))))..)))(((((((((.......))).)))))).))).((((((((((((((((((.......))))))....))))))))))..)).. ( -42.20) >DroVir_CAF1 61299 118 + 1 GGUCAU--UUUGUAGGUACUGGCAAGACUGCCACCUUCUCGAUUGCCAUCCUUCAGCAAAUCGACACGUCCAUGCGCGAAUGUCAGGCUUUGAUCCUGGCACCAACUCGCGAAUUGGCCA (((((.--((((((((.(((((((....))))).....((((((((.........)).))))))...)))).)))((((.(((((((.......))))))).....))))))).))))). ( -37.00) >DroGri_CAF1 49563 120 + 1 GUUUGUUUGUUUUAGGUACUGGCAAAACUGCCACCUUUUCGAUUGCCAUCCUUCAGCAAAUCGAUACGGCCGUUCGUGAAUGCCAGGCUCUGAUCUUGGCCCCCACUCGCGAGUUGGCUA (((.(((((((..(((...((((((.................)))))).)))..))))))).)))..((((..((((((.((...((((........))))..)).))))))...)))). ( -34.13) >DroWil_CAF1 66290 117 + 1 UAU-A--UUUAACAGGUACUGGCAAAACUGCCACCUUCUCGAUUGCCAUUCUGCAGCAAAUCGACACGACAAGUCGCGAAUGUCAGGCUCUGAUUUUGGCACCAACUCGUGAAUUGGCCA ...-.--.......(((..(((((....))))).....((((((((.........)).))))))(((((...((.((.((.(((((...))))).)).))))....))))).....))). ( -28.50) >DroMoj_CAF1 57771 119 + 1 GCU-AUCGCUUGUAGGUACUGGCAAGACUGCCACCUUCUCGAUUGCCAUACUUCAGCAAAUCGACACGUCCAUUCGCGAAUGCCAAGCUUUGAUCUUGGCACCCACUCGCGAGUUGGCCA ...-...(((.(.(((((.((((((.................)))))))))))))))..........(((.((((((((.(((((((.......))))))).....)))))))).))).. ( -35.83) >DroAna_CAF1 41629 119 + 1 AUU-UACUUUCACAGGAACUGGCAAGACUGCCACCUUCUCGAUCGCCAUCCUUCAGCAGAUCGACACGUCCAUUCGCGAGUGCCAGGCUCUGAUCCUGGCCCCAACUCGCGAGCUGGCCA ...-..........(((..(((((....)))))..)))((((((((.........)).))))))...(.((((((((((((((((((.......))))))....))))))))).))).). ( -42.30) >consensus GAU_AUCUUUUGUAGGUACUGGCAAGACUGCCACCUUCUCGAUUGCCAUCCUUCAGCAAAUCGACACGUCCAUUCGCGAAUGCCAGGCUCUGAUCCUGGCACCAACUCGCGAGUUGGCCA .............(((...(((((....))))))))..((((((((.........)).))))))...(.((((((((((..((((((.......))))))......))))))).))).). (-29.87 = -28.85 + -1.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:23 2006