
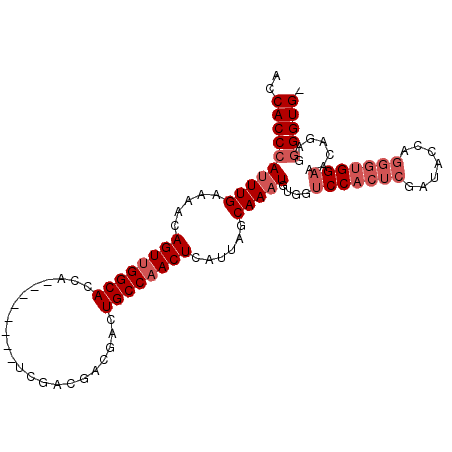
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,977,335 – 5,977,430 |
| Length | 95 |
| Max. P | 0.758808 |

| Location | 5,977,335 – 5,977,430 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 84.90 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -25.00 |
| Energy contribution | -26.33 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
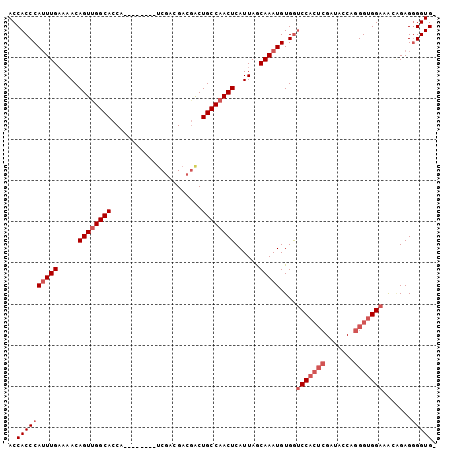
>2L_DroMel_CAF1 5977335 95 + 22407834 ACCACCCAUUUGAAAACAGUAGGCACCA--------UCGACGACGACUGCCAACUCAUUAGCAAAUGUGGUCCACUCGAUACCGGGGUGGAAACAGAGGGGUG- ..((((((((((.....(((.((((...--------(((....))).)))).)))......)))))((..(((((((.......))))))).))....)))))- ( -30.60) >DroSec_CAF1 33395 95 + 1 ACCACCCAUUUGAAAACAGUUGGCACCA--------UCGACGACGACUGCCAACUCAUUAGCAAAUGUGGUCCACUCGAUCCCAGGGUGGAAACAGAGGGGUG- ..((((((((((.....((((((((...--------(((....))).))))))))......)))))((..(((((((.......))))))).))....)))))- ( -35.30) >DroSim_CAF1 34093 95 + 1 ACCACCCAUUUGAAAACAGUUGGCACCA--------UCGACGACGACUGCCAACUCAUUAGCAAAUGUGGUCCACUCGAUACCAGGGUGGAAACAGAGGGGUG- ..((((((((((.....((((((((...--------(((....))).))))))))......)))))((..(((((((.......))))))).))....)))))- ( -35.30) >DroEre_CAF1 38254 95 + 1 ACCACCCAUUUGAAAACAGUUGGCACCA--------UCGACGACGGCUGCCAACUCAUUAGCAAAUGUGGUCCACUCGAUGCCAGGGUGGAAAAGGAGGGGUG- (((.((((((((.....((((((((((.--------((...)).)).))))))))......)))))).))(((((((.......)))))))........))).- ( -34.90) >DroYak_CAF1 37474 95 + 1 ACCACCCAUUUGAAAACAGUUGGCACCA--------UCGACGACGACUGCCAACUCAUUAGCAAAUGUGGUCCACUCGAUUCCGGGGUGGAAAAGGAGGGGUG- (((.((((((((.....((((((((...--------(((....))).))))))))......)))))).))(((((((.......)))))))........))).- ( -33.20) >DroAna_CAF1 35399 96 + 1 ACCACCCAAUUGCAAACAGUUGGCAACAAAAUGCGGCCUCCGCCAACUGCCAACUCAUUAACAAAUGUGAGCCAGGA--------GGAGGAGGGUGAGAGGUGG .((((((..(((((.....(((....)))..)))))(((((.((..(((....(((((........))))).)))..--------)).)))))....).))))) ( -34.60) >consensus ACCACCCAUUUGAAAACAGUUGGCACCA________UCGACGACGACUGCCAACUCAUUAGCAAAUGUGGUCCACUCGAUACCAGGGUGGAAACAGAGGGGUG_ ..((((((((((.....((((((((......................))))))))......)))))....(((((((.......))))))).......))))). (-25.00 = -26.33 + 1.33)

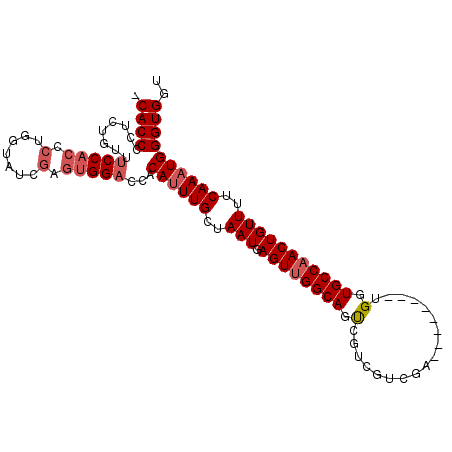
| Location | 5,977,335 – 5,977,430 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 84.90 |
| Mean single sequence MFE | -32.93 |
| Consensus MFE | -23.94 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
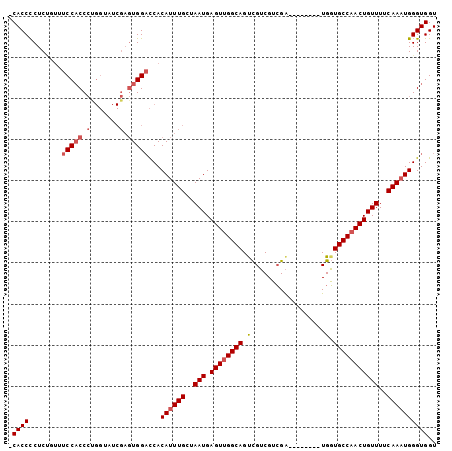
>2L_DroMel_CAF1 5977335 95 - 22407834 -CACCCCUCUGUUUCCACCCCGGUAUCGAGUGGACCACAUUUGCUAAUGAGUUGGCAGUCGUCGUCGA--------UGGUGCCUACUGUUUUCAAAUGGGUGGU -((((........(((((..((....)).)))))...((((((..(((.(((.((((.(((((...))--------))))))).))))))..)))))))))).. ( -29.50) >DroSec_CAF1 33395 95 - 1 -CACCCCUCUGUUUCCACCCUGGGAUCGAGUGGACCACAUUUGCUAAUGAGUUGGCAGUCGUCGUCGA--------UGGUGCCAACUGUUUUCAAAUGGGUGGU -..((.(((...((((.....))))..))).))(((.((((((..(((.((((((((.(((((...))--------))))))))))))))..)))))))))... ( -34.70) >DroSim_CAF1 34093 95 - 1 -CACCCCUCUGUUUCCACCCUGGUAUCGAGUGGACCACAUUUGCUAAUGAGUUGGCAGUCGUCGUCGA--------UGGUGCCAACUGUUUUCAAAUGGGUGGU -..((.(((.((..((.....)).)).))).))(((.((((((..(((.((((((((.(((((...))--------))))))))))))))..)))))))))... ( -32.50) >DroEre_CAF1 38254 95 - 1 -CACCCCUCCUUUUCCACCCUGGCAUCGAGUGGACCACAUUUGCUAAUGAGUUGGCAGCCGUCGUCGA--------UGGUGCCAACUGUUUUCAAAUGGGUGGU -((((........(((((.(.......).)))))...((((((..(((.((((((((.(((((...))--------))))))))))))))..)))))))))).. ( -35.10) >DroYak_CAF1 37474 95 - 1 -CACCCCUCCUUUUCCACCCCGGAAUCGAGUGGACCACAUUUGCUAAUGAGUUGGCAGUCGUCGUCGA--------UGGUGCCAACUGUUUUCAAAUGGGUGGU -..((.(((...((((.....))))..))).))(((.((((((..(((.((((((((.(((((...))--------))))))))))))))..)))))))))... ( -34.90) >DroAna_CAF1 35399 96 - 1 CCACCUCUCACCCUCCUCC--------UCCUGGCUCACAUUUGUUAAUGAGUUGGCAGUUGGCGGAGGCCGCAUUUUGUUGCCAACUGUUUGCAAUUGGGUGGU (((((............((--------....))....((.((((.(((.(((((((((...((((...))))......)))))))))))).)))).))))))). ( -30.90) >consensus _CACCCCUCUGUUUCCACCCUGGUAUCGAGUGGACCACAUUUGCUAAUGAGUUGGCAGUCGUCGUCGA________UGGUGCCAACUGUUUUCAAAUGGGUGGU .((((........(((((.(.......).)))))...((((((..(((.((((((((.(..................).)))))))))))..)))))))))).. (-23.94 = -24.80 + 0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:19 2006