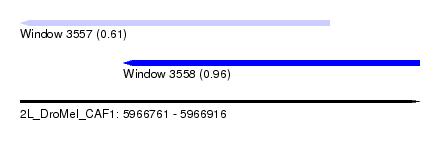
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,966,761 – 5,966,916 |
| Length | 155 |
| Max. P | 0.964240 |

| Location | 5,966,761 – 5,966,881 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.78 |
| Mean single sequence MFE | -46.13 |
| Consensus MFE | -13.87 |
| Energy contribution | -13.73 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5966761 120 - 22407834 AGCUGGCAUGCAAGAAUUUCAUCACCCAAUUGUGGGCAGCCCGCCAGCAGCUCCACCAGGAGUUCCAGCUGGCCCAAGCCCAGCGUCCACGGCCGCUUCGUAUGAUCCUGCAGCAUUUGC .((..((.((((.((...((((.((.......(((((.....((((((((((((....))))))...))))))....)))))(((.(....).)))...)))))))).))))))....)) ( -41.80) >DroVir_CAF1 37765 120 - 1 AUAUGGCGUGCCAGCAGUUUCUUACUCAGCUGCAGGCGGCGCAGCAGCAGCUGCACGCGGAACUGCAGCAGGCUCAGGCCCAGCGACCACGCCCAAUUCAUAUGCUGCUGCAGCGCCUAC ....((((((((.((((((........)))))).)))...(((((((((((((((........)))))).(((....)))..(((....)))..........))))))))).)))))... ( -55.30) >DroPse_CAF1 40593 120 - 1 AUAUGGCCUGCCAGCAGUUCCUCGCCCAAAUCCAGGCAGCCCGCCAGCACUUCAAUACGGAGCUCCUGCAGGCCCAGGCCCAACGGCCACGACCACUGCAUAUAAUCCUGCAGCAGCUCA ....(((((((.((.((((((..(((........))).((......))..........)))))).)))))))))..((((....))))..(((..(((((........)))))..).)). ( -43.90) >DroWil_CAF1 40917 120 - 1 GUAUGGCAUGCCAACAGUUCCUUACCCAAAUCAAGGAGGCCCAUCAGCAGUUGCAGGAGGAAUUGCAGCAGGCCCAGGCCCAGCGGCCACGCCCAUUACAUAUAAUCCUGCAGCAGCUAA (.((((...(((......(((((.........)))))))))))))(((.(((((((((.....((.((..(((...((((....))))..)))..)).)).....))))))))).))).. ( -38.90) >DroMoj_CAF1 34237 120 - 1 AUAUGGCAUGCCAGCAGUUCAUUACCCAGGUGCAGGCGGCGCACCAGCAGCUGCACGUGGAAAUGCAGCAGGCGCACGCCCAGCGCCCGCGGCCGCUGCAUAUGCUGAUGCAGCAGCUCC ....(((..(((.(((((.....))(((.((((((.(.((......)).))))))).)))...))).((.(((((.......))))).))))).(((((((......))))))).))).. ( -52.80) >DroAna_CAF1 25356 120 - 1 GUAUGGCCUGCCAGCAGUUCCUCACCCACCUCUUGGCGGCCCAACAGCAGUUUCACCAGGAGUUCCAGUUGGCCCAGGCCCAACGGCCACGGCCUCUCCGCAUGAUCCUGCAGCGGCUGC ...(((....)))(((((..((...((.((....)).)).......((((..(((...((((........((((..((((....))))..)))).))))...)))..))))))..))))) ( -44.10) >consensus AUAUGGCAUGCCAGCAGUUCCUCACCCAAAUCCAGGCGGCCCACCAGCAGCUGCACGAGGAAUUCCAGCAGGCCCAGGCCCAGCGGCCACGGCCACUGCAUAUGAUCCUGCAGCAGCUAC ....(((.(((....((((((...((........))..((......))..........))))))...((.(((....)))...((....))..................)).)))))).. (-13.87 = -13.73 + -0.13)

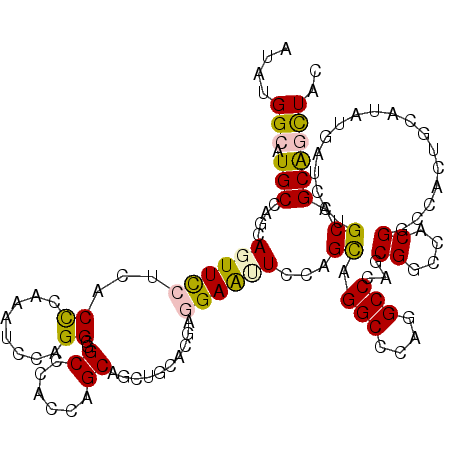

| Location | 5,966,801 – 5,966,916 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 73.26 |
| Mean single sequence MFE | -45.28 |
| Consensus MFE | -13.66 |
| Energy contribution | -13.83 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.30 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5966801 115 - 22407834 AUG--GCAGCCCCUCCAACUGCACCUGCAGCGGAGUCAGCUGGCAUGCAAGAAUUUCAUCACCCAAUUGUGGGCAGCCCGCCAGCAGCUCCACCAGGAGUUCCAGCUGGCCCAAGCC (((--.((((..((((..((((....)))).))))...)))).)))((.............((((....))))......((((((((((((....))))))...))))))....)). ( -44.40) >DroVir_CAF1 37805 115 - 1 CCU--GCUGCCUUUGCAGCUGGGCCUGCUCCGCAAUCAUAUGGCGUGCCAGCAGUUUCUUACUCAGCUGCAGGCGGCGCAGCAGCAGCUGCACGCGGAACUGCAGCAGGCUCAGGCC (((--(((((....))))).(((((((((((((.........((.((((.((((((........)))))).))))))(((((....)))))..)))))......))))))))))).. ( -57.20) >DroPse_CAF1 40633 115 - 1 AUG--GCUCCCACUCCGGCUGCAUCUCCUGCGCAGUCAUAUGGCCUGCCAGCAGUUCCUCGCCCAAAUCCAGGCAGCCCGCCAGCACUUCAAUACGGAGCUCCUGCAGGCCCAGGCC ..(--(((........((((((.........))))))....(((((((.((.((((((..(((........))).((......))..........)))))).)))))))))..)))) ( -43.30) >DroWil_CAF1 40957 117 - 1 AUCCUUCUCCCACUGCGCCUGCUGCUCCUACGCAAUCGUAUGGCAUGCCAACAGUUCCUUACCCAAAUCAAGGAGGCCCAUCAGCAGUUGCAGGAGGAAUUGCAGCAGGCCCAGGCC ............(((.(((((((((((((..(((((.((((((...(((......(((((.........))))))))))))..)).)))))...))))...))))))))).)))... ( -42.70) >DroAna_CAF1 25396 115 - 1 AUG--GCUGCCCCUGCAUUUUCACCUCCGGCGCAGCCGUAUGGCCUGCCAGCAGUUCCUCACCCACCUCUUGGCGGCCCAACAGCAGUUUCACCAGGAGUUCCAGUUGGCCCAGGCC (((--(((((.((...............)).))))))))..(((((((((((.(.....)......(((((((.((..(.......)..)).))))))).....))))))..))))) ( -40.76) >DroPer_CAF1 40393 115 - 1 AUG--GCUCCCACUCCGGCUGCAUCUCCUGCGCAGUCAUAUGGCCUGCCAGCAGUUCCUCGCCCAAAUCCAGGCAGCCCGCCAGCACUUCAAUACGGAGCUCCUGCAGGCCCAGGCC ..(--(((........((((((.........))))))....(((((((.((.((((((..(((........))).((......))..........)))))).)))))))))..)))) ( -43.30) >consensus AUG__GCUCCCACUCCAGCUGCACCUCCUGCGCAGUCAUAUGGCCUGCCAGCAGUUCCUCACCCAAAUCCAGGCAGCCCACCAGCAGUUCCACAAGGAGCUCCAGCAGGCCCAGGCC .....(((....((((...............))))......(((.(((....((((((...((........))..((......))..........))))))...))).)))..))). (-13.66 = -13.83 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:15 2006