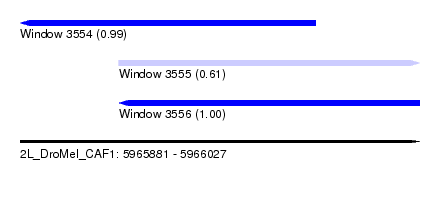
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,965,881 – 5,966,027 |
| Length | 146 |
| Max. P | 0.995441 |

| Location | 5,965,881 – 5,965,989 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.23 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -12.74 |
| Energy contribution | -13.05 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
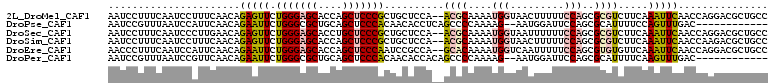
>2L_DroMel_CAF1 5965881 108 - 22407834 AAUCCUUUCAAUCCUUUCAACAGAGUUCUGGGAGCACCAGCUCCCGCUGCUCCA--ACGCAAAAUGGUAACUUUUUCCAGCGCGUCUUCAAAUUCAACCAGGACGCUGCC ......................((((..(((((((....)))))))..))))..--..(((...(((.........)))..(((((((...........)))))))))). ( -29.80) >DroPse_CAF1 39260 96 - 1 AAUCCGUUUAAUCCAUUCAACAGAAUUCUGGGCGCUGCAGCUCCCACAACACCUCAGCCCCAAAAG--AAUGGAUUCCAGCGCAUUUUCCAGUUUGAC------------ ....((((.(((((((((..(((....)))((.((((.((............)))))).))....)--))))))))..))))................------------ ( -23.30) >DroSec_CAF1 21911 108 - 1 AAUCCUUUCAAUCCCUUGAACAGAGUUCUGGGAGCACCUGCUCCCGCUGCUCCA--ACGCAAAAUGGUAAUUUUUUCCAGCGCGUCUUCAAAUUCAACCAGGACGCUGCC ......(((((....)))))..((((..(((((((....)))))))..))))..--..(((...(((.........)))..(((((((...........)))))))))). ( -31.30) >DroSim_CAF1 22028 108 - 1 AAUCCUUUCAAUCCUUUCAACAGAGUUCUGGGAGCACCAGCUCCCGCUGCUCCA--ACGCAAAAUGGUAACUUUUUCCAGCGCGUCUUCAAAUUCAACCAAGACGCUGCC ......................((((..(((((((....)))))))..))))..--..(((...(((.........)))..(((((((...........)))))))))). ( -30.10) >DroEre_CAF1 26588 108 - 1 AACCCUUUCAAUCCAUUCAACAGAAUUCUGGGAGCACCAGCUCCCAAUCCGCCA--GCACAAAAUGGUCAAUUUUUCCAGCGUGUGUUCAAAUUCAACCAGGACGCUGCC ....................(((..((((((((((....))))))........(--(((((...(((.........)))...))))))...........))))..))).. ( -26.20) >DroPer_CAF1 38990 96 - 1 AAUCCGUUUAAUCCGUUCAACAGAAUUCUGGGCGCUGCAGCUCCCACAACACCACAGCCCCAAAAG--AAUGGAUUCCAGCGCAUUUUCAAGUUUGAC------------ ....((((.(((((((((..(((....)))((.((((.................)))).))....)--))))))))..))))................------------ ( -22.23) >consensus AAUCCUUUCAAUCCAUUCAACAGAAUUCUGGGAGCACCAGCUCCCACUGCUCCA__ACGCAAAAUGGUAAUUUUUUCCAGCGCGUCUUCAAAUUCAACCAGGACGCUGCC ......................(((((.(((((((....)))))))..........((((....(((.........)))..)))).....)))))............... (-12.74 = -13.05 + 0.31)
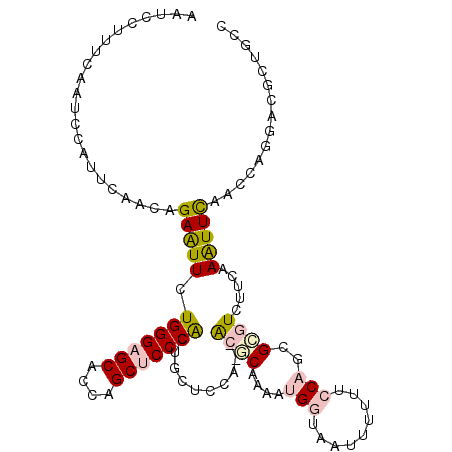
| Location | 5,965,917 – 5,966,027 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.16 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -19.61 |
| Energy contribution | -20.45 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5965917 110 + 22407834 AAAGUUACCAUUUUGCGU--UGGAGCAGCGGGAGCUGGUGCUCCCAGAACUCUGUUGAAAGGAUUGAAAGGAUUGAGGGCUCCGAAGGGUCCGAAUGGGUUCAUUCCCAUUC .......((.(((..(.(--(....((((((((((....))))))((....)))))).)).)...))).)).....((((((....))))))(((((((......))))))) ( -36.90) >DroPse_CAF1 39284 110 + 1 UCCAUU--CUUUUGGGGCUGAGGUGUUGUGGGAGCUGCAGCGCCCAGAAUUCUGUUGAAUGGAUUAAACGGAUUCAGGCUUCCGAAUGGGCCAAACGGGUUGAGACCCAUUC ((((((--(...((((.(((.(((((((..(...)..))))))))))...))))..)))))))......(((........)))(((((((((((.....))).).))))))) ( -39.90) >DroSim_CAF1 22064 110 + 1 AAAGUUACCAUUUUGCGU--UGGAGCAGCGGGAGCUGGUGCUCCCAGAACUCUGUUGAAAGGAUUGAAAGGAUUGAGGGCUCCGAAGGGUCCGAAUGGGUUCAUUCCCAUUC .......((.(((..(.(--(....((((((((((....))))))((....)))))).)).)...))).)).....((((((....))))))(((((((......))))))) ( -36.90) >DroEre_CAF1 26624 110 + 1 AAAUUGACCAUUUUGUGC--UGGCGGAUUGGGAGCUGGUGCUCCCAGAAUUCUGUUGAAUGGAUUGAAAGGGUUCAGGGCUCCGAAAGGUCCGAAUGGGCUCAUUCCCAUUC .....((((.(((((.((--(((((((((((((((....))))))))...))))))...((((((.....)))))).)))..))))))))).(((((((......))))))) ( -40.60) >DroWil_CAF1 39914 105 + 1 AGGAUU--CAUGGUUGGUUGAGGUGUGGUUGGAGCUGGAGCUCCCAGUAUCCGAUUGAAGGGAUUGAAGGGAUUUAGAUUUCCCAAAGCUCCGAAUGGAUUCAAUAC----- .(((((--(((..(..((((.((((..(..(((((....))))))..))))))))..).((..((...((((........))))..))..))..)))))))).....----- ( -32.00) >DroPer_CAF1 39014 110 + 1 UCCAUU--CUUUUGGGGCUGUGGUGUUGUGGGAGCUGCAGCGCCCAGAAUUCUGUUGAACGGAUUAAACGGAUUCAGGCUUCCGAAUGGGCCAAACGGGCUCAGACCCAUUC ......--....((((.(((.(((((((..(...)..))))))))))...((((.....)))).....((((........))))..((((((.....))))))..))))... ( -38.00) >consensus AAAAUU__CAUUUUGGGC__UGGUGUAGUGGGAGCUGGAGCUCCCAGAAUUCUGUUGAAAGGAUUGAAAGGAUUCAGGGCUCCGAAAGGUCCGAAUGGGUUCAUUCCCAUUC .....................((......((((((....)))))).((((((.(((....(((((....(((........)))....))))).)))))))))....)).... (-19.61 = -20.45 + 0.84)

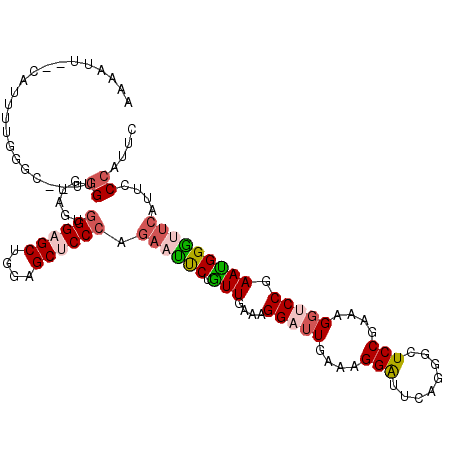
| Location | 5,965,917 – 5,966,027 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.16 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -19.50 |
| Energy contribution | -20.62 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5965917 110 - 22407834 GAAUGGGAAUGAACCCAUUCGGACCCUUCGGAGCCCUCAAUCCUUUCAAUCCUUUCAACAGAGUUCUGGGAGCACCAGCUCCCGCUGCUCCA--ACGCAAAAUGGUAACUUU (((((((......)))))))(((......(((........)))......)))........((((..(((((((....)))))))..))))..--.................. ( -33.70) >DroPse_CAF1 39284 110 - 1 GAAUGGGUCUCAACCCGUUUGGCCCAUUCGGAAGCCUGAAUCCGUUUAAUCCAUUCAACAGAAUUCUGGGCGCUGCAGCUCCCACAACACCUCAGCCCCAAAAG--AAUGGA (((((((((...........)))))))))(((........)))......(((((((..(((....)))((.((((.((............)))))).))....)--)))))) ( -35.20) >DroSim_CAF1 22064 110 - 1 GAAUGGGAAUGAACCCAUUCGGACCCUUCGGAGCCCUCAAUCCUUUCAAUCCUUUCAACAGAGUUCUGGGAGCACCAGCUCCCGCUGCUCCA--ACGCAAAAUGGUAACUUU (((((((......)))))))(((......(((........)))......)))........((((..(((((((....)))))))..))))..--.................. ( -33.70) >DroEre_CAF1 26624 110 - 1 GAAUGGGAAUGAGCCCAUUCGGACCUUUCGGAGCCCUGAACCCUUUCAAUCCAUUCAACAGAAUUCUGGGAGCACCAGCUCCCAAUCCGCCA--GCACAAAAUGGUCAAUUU (((((((......)))))))(((......(((....((((....)))).)))..............(((((((....))))))).)))((((--........))))...... ( -34.40) >DroWil_CAF1 39914 105 - 1 -----GUAUUGAAUCCAUUCGGAGCUUUGGGAAAUCUAAAUCCCUUCAAUCCCUUCAAUCGGAUACUGGGAGCUCCAGCUCCAACCACACCUCAACCAACCAUG--AAUCCU -----(.(((((((((....))).....((((........)))).........)))))))((((....(((((....))))).........(((........))--))))). ( -23.20) >DroPer_CAF1 39014 110 - 1 GAAUGGGUCUGAGCCCGUUUGGCCCAUUCGGAAGCCUGAAUCCGUUUAAUCCGUUCAACAGAAUUCUGGGCGCUGCAGCUCCCACAACACCACAGCCCCAAAAG--AAUGGA (((((((.(..((....))..))))))))(((........)))......(((((((..(((....)))((.((((.................)))).))....)--)))))) ( -34.33) >consensus GAAUGGGAAUGAACCCAUUCGGACCAUUCGGAACCCUGAAUCCUUUCAAUCCAUUCAACAGAAUUCUGGGAGCACCAGCUCCCACUACACCA__ACCCAAAAUG__AAUUUU (((((((......)))))))((.......(((........))).......................(((((((....))))))).....))..................... (-19.50 = -20.62 + 1.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:13 2006