| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 631,158 – 631,248 |
| Length | 90 |
| Max. P | 0.695766 |

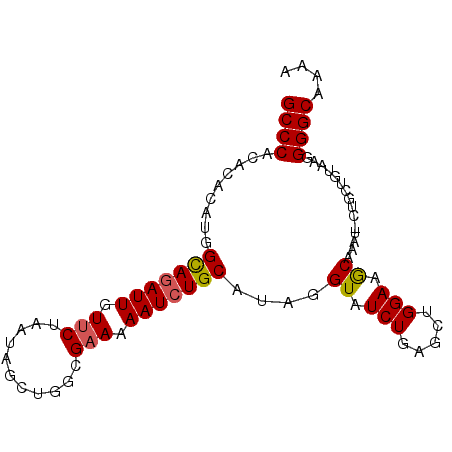
| Location | 631,158 – 631,248 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 91.07 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 631158 90 + 22407834 UUUUGCCCCUUACUGCAGUAUUUGCUUCCAGCUAAGAUACCUAUGCAGAUUUUUCGCCAGCUAUUAGAACAAUCUACCAUGUGUGUGGGC ....(((((((((((.(((....)))..))).))))((((.((((.(((((.(((...........))).)))))..)))).)))))))) ( -20.30) >DroSec_CAF1 20087 90 + 1 UUUUGCCCCUUACAGCAGGAUUUGCUUCCAGCUCAGAUACCUAUGCAGAUUUUGCGCCAGCUAUUAGAACAAUUUGCCAUGUGUGUGGGC ....(((((.(((((((((((((((...................)))))))))))....((..............))..)))).).)))) ( -22.35) >DroSim_CAF1 19836 90 + 1 UUUUGCCCCUUACAGCAGGAUUUGCUUCCAGCUCAGAUACCUAUGCAGAUUUUUCGCCAGCUAUUAGAACAAUCUGCCAUGUGUGUGGGC ....((((.....(((.(((......))).)))...((((.((((((((((.(((...........))).)))))).)))).)))))))) ( -25.60) >DroEre_CAF1 24117 86 + 1 UUUAGCCCCUUGCAAUA----UUGUUUCCAGCUCAGAUACCUAUGCAGAUUUUUCGCCAGCCAUUAGAACAAUCUGCCAUGUGUGUGGGC ....((.....))....----.........(((((.((((....(((((((.(((...........))).)))))))...)))).))))) ( -21.30) >DroYak_CAF1 23242 86 + 1 UUUAGCCCCUUACAAUA----UUGUUUCCAGCUCAGAUACCUAUGCAGAUUUUUCGCCAGCCAUUAGAACAAUCUGCCAUGUGUGUGGGC .................----.........(((((.((((....(((((((.(((...........))).)))))))...)))).))))) ( -20.80) >consensus UUUUGCCCCUUACAGCAG_AUUUGCUUCCAGCUCAGAUACCUAUGCAGAUUUUUCGCCAGCUAUUAGAACAAUCUGCCAUGUGUGUGGGC ....((((...............((.....))....((((.((((((((((.(((...........))).)))))).)))).)))))))) (-17.60 = -17.60 + -0.00)

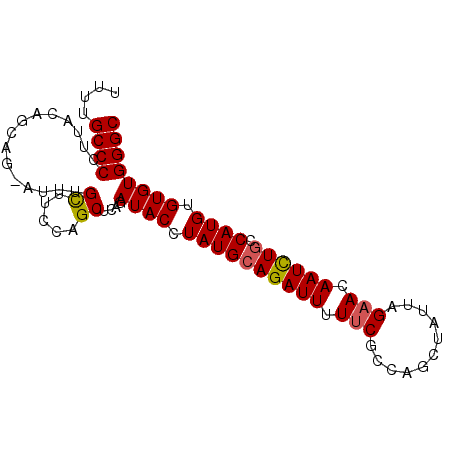
| Location | 631,158 – 631,248 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 91.07 |
| Mean single sequence MFE | -23.64 |
| Consensus MFE | -18.74 |
| Energy contribution | -18.74 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 631158 90 - 22407834 GCCCACACACAUGGUAGAUUGUUCUAAUAGCUGGCGAAAAAUCUGCAUAGGUAUCUUAGCUGGAAGCAAAUACUGCAGUAAGGGGCAAAA ((((....((...(((((((.(((...........))).)))))))....))..((((.(((..((......)).))))))))))).... ( -21.90) >DroSec_CAF1 20087 90 - 1 GCCCACACACAUGGCAAAUUGUUCUAAUAGCUGGCGCAAAAUCUGCAUAGGUAUCUGAGCUGGAAGCAAAUCCUGCUGUAAGGGGCAAAA ((((......((((((....((((..(((.(((..(((.....))).))).)))..)))).(((......)))))))))...)))).... ( -26.00) >DroSim_CAF1 19836 90 - 1 GCCCACACACAUGGCAGAUUGUUCUAAUAGCUGGCGAAAAAUCUGCAUAGGUAUCUGAGCUGGAAGCAAAUCCUGCUGUAAGGGGCAAAA ((((..((((...(((((((.(((...........))).)))))))....)).....(((.(((......))).)))))...)))).... ( -26.00) >DroEre_CAF1 24117 86 - 1 GCCCACACACAUGGCAGAUUGUUCUAAUGGCUGGCGAAAAAUCUGCAUAGGUAUCUGAGCUGGAAACAA----UAUUGCAAGGGGCUAAA ((((.((....))((((...((((..(((.((((((.......))).))).)))..))))((....)).----..))))...)))).... ( -22.70) >DroYak_CAF1 23242 86 - 1 GCCCACACACAUGGCAGAUUGUUCUAAUGGCUGGCGAAAAAUCUGCAUAGGUAUCUGAGCUGGAAACAA----UAUUGUAAGGGGCUAAA ((((..(((....(((((((.(((...........))).)))))))..............((....)).----...)))...)))).... ( -21.60) >consensus GCCCACACACAUGGCAGAUUGUUCUAAUAGCUGGCGAAAAAUCUGCAUAGGUAUCUGAGCUGGAAGCAAAU_CUGCUGUAAGGGGCAAAA ((((.........(((((((.(((...........))).)))))))....((.(((.....))).))...............)))).... (-18.74 = -18.74 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:26 2006