
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,941,588 – 5,941,717 |
| Length | 129 |
| Max. P | 0.550573 |

| Location | 5,941,588 – 5,941,678 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -19.59 |
| Energy contribution | -19.52 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5941588 90 + 22407834 GUGGGCCGGCAU-AUCAUGCUGCAGGAUUGAUAGCUGUUUGCUAAUUUUGAGUUUUCCGCAUACGCCUAGUAUGACGGGUACUCCAGGUCA ...((((((((.-..((.(((((......).))))))..))))......((((..(((((((((.....))))).)))).))))..)))). ( -29.70) >DroSec_CAF1 4230 85 + 1 GUGGGCCGGCAUUAUCAUGCUGCAGGAUUGAUAGCUGUUUGCUAAU-UUGAGUUUUCCGCAUACGCCUAGUAUGACGGGUACUCC-----A ...(((..(((((((((..(.....)..)))))).)))..)))...-..((((..(((((((((.....))))).)))).)))).-----. ( -25.10) >DroEre_CAF1 4305 89 + 1 GUGGGCCGGCAU-AUCAUGCUGCAGGAUUGAUAGCUGUUUGCUAAUUUUGAGU-UGCUGCAUACGCCUGGUAUGACGGAUAUACCAGUUCG .(((.(((.(((-((((.(((((((((((.(((((.....))))....).)))-).)))))...)).))))))).))).....)))..... ( -27.20) >DroYak_CAF1 3976 89 + 1 GUGGGCCGGCAU-AUCAUGCUGCAGGAUUGAUAGCUGUUUGCUAAUUUUGAGU-UUCUGCAUGCGCCUAGUAUGACGGAAAUAACAGGUCA ...(((((((((-...))))).((((((...((((.....)))))))))).((-((((((((((.....))))).)))))))....)))). ( -27.30) >consensus GUGGGCCGGCAU_AUCAUGCUGCAGGAUUGAUAGCUGUUUGCUAAUUUUGAGU_UUCCGCAUACGCCUAGUAUGACGGAUACACCAGGUCA ......((((((....))))))((((((...((((.....)))))))))).....(((((((((.....))))).))))............ (-19.59 = -19.52 + -0.06)

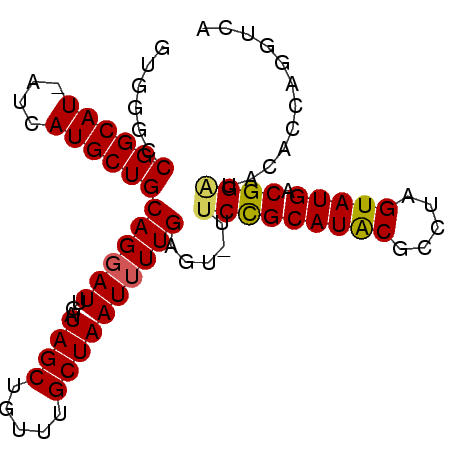

| Location | 5,941,615 – 5,941,717 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.22 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -22.85 |
| Energy contribution | -23.23 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5941615 102 + 22407834 UGAUAGCUGUUUGCUAAUUUUGAGUUUUCCGCAUACGCCUAGUAUGACGGGUACUCCAGGUCAGCGGGUGUGUAUGGAAA-GACCACGUAUAUGGCUGUAAUC ..((((((..(((((.((((.((((..(((((((((.....))))).)))).)))).)))).)))))(((((..(((...-..)))..))))))))))).... ( -34.30) >DroSec_CAF1 4258 96 + 1 UGAUAGCUGUUUGCUAAU-UUGAGUUUUCCGCAUACGCCUAGUAUGACGGGUACUCC-----AGCGGGUGUGUAUGGAAA-GACCACGUGUAUGGCUGUAAUC ..((((((((........-..((((..(((((((((.....))))).)))).)))).-----.(((.(((.((.(....)-.))))).))))))))))).... ( -28.80) >DroEre_CAF1 4332 100 + 1 UGAUAGCUGUUUGCUAAUUUUGAGU-UGCUGCAUACGCCUGGUAUGACGGAUAUACCAGUUCGGCGGAUGUGGUGGGAA--CUCCACGUAUAUGGUUGUAAUC ...((((.....)))).......((-(((..(((((((((((((((.....)))))))....)))).((((((.(....--).)))))).))))...))))). ( -29.70) >DroYak_CAF1 4003 102 + 1 UGAUAGCUGUUUGCUAAUUUUGAGU-UUCUGCAUGCGCCUAGUAUGACGGAAAUAACAGGUCAGCGGCUGCGAUGGGAAACAUCCGCGUAUAUGGCUGUAAUC ..((((((((.(((.........((-((((((((((.....))))).)))))))....(((.....)))(((...(....)...)))))).)))))))).... ( -30.30) >consensus UGAUAGCUGUUUGCUAAUUUUGAGU_UUCCGCAUACGCCUAGUAUGACGGAUACACCAGGUCAGCGGGUGUGUAGGGAAA_GACCACGUAUAUGGCUGUAAUC ..(((((((((((((.((((.((((...((((((((.....))))).)))..)))).)))).)))))..(((............)))....)))))))).... (-22.85 = -23.23 + 0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:04 2006