
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,936,114 – 5,936,249 |
| Length | 135 |
| Max. P | 0.758194 |

| Location | 5,936,114 – 5,936,210 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.58 |
| Mean single sequence MFE | -19.21 |
| Consensus MFE | -14.72 |
| Energy contribution | -14.22 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
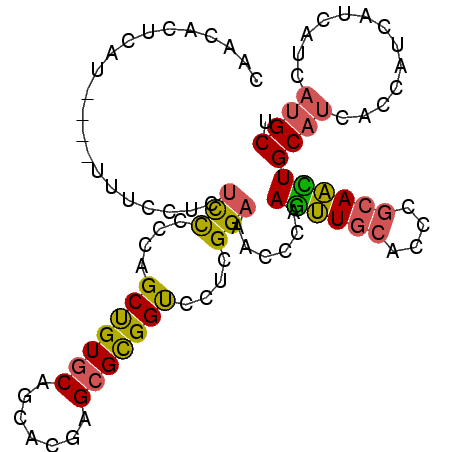
>2L_DroMel_CAF1 5936114 96 + 22407834 CAACACUCAU----UUUCCUCUCCCCCAGCUGUGCAGCACGAGCGCGGUCCUAGGAAACCCAAGUUGCACCCGCAACUGCAUCAUCAUCAUCAUCAUGCU ..........----((((((........(((((((.......)))))))...))))))....((((((....))))))((((.((.......)).)))). ( -20.70) >DroPse_CAF1 27165 98 + 1 CCUCUAUCUC--UCUCUCUCUUCUUACAGCCGUUCAACACGAGCGUGGUCCUCGGAAACCCAAGCUCCAUCCGCAGUUGCAUCACCAUCAUCAUCACGCG ..........--..................(((.....))).(((((((....(((.........)))....((....))............))))))). ( -14.10) >DroSec_CAF1 21972 95 + 1 CAACACUCAU----UUUCCUCUCC-CCAGCUGUGCAGCACGAGCGCGGCCCCAGGAAACCGAAGUUGCACCCGCAACUGCAUCACCAUCAUCAUCAUGCU ......((..----((((((....-...(((((((.......)))))))...))))))..))((((((....))))))((((.............)))). ( -23.12) >DroEre_CAF1 22043 96 + 1 CAACACCCAU----UUUCCUCUCCCCCAGCUGUGCAGCACGAGCGCGGUCCACGGAAACCCAAGUUGCACCCGCAACUGCAUCACCAUCACCAUCAUGCU ..........----.......(((....(((((((.......)))))))....)))......((((((....))))))((((.............)))). ( -20.22) >DroWil_CAF1 32074 91 + 1 -------AUUUCU-UAUCUC-AUUCUCAGCUGUGCAGCACGAGCGUGGUCCUCGGAAACCGAAAUUGCAUCCCCAAUUGCAUCAUCAUCAUCAUCAUGCG -------......-......-.......(((....)))....(((((((..((((...))))...((((........))))...........))))))). ( -17.60) >DroYak_CAF1 22184 96 + 1 CAACACCCAU----UCUCCUCUCCCCCAGCUGUGCAGCACGAGCGCGGUCCACGGAAACCGAAGUUGCAUCCGCAACUGCACCACCAUCAUCAUCAUGCU ..........----.......(((....(((((((.......)))))))....)))....(.((((((....)))))).).................... ( -19.50) >consensus CAACACUCAU____UUUCCUCUCCCCCAGCUGUGCAGCACGAGCGCGGUCCUCGGAAACCCAAGUUGCACCCGCAACUGCAUCACCAUCAUCAUCAUGCU .....................(((....(((((((.......)))))))....)))......((((((....))))))((((.............)))). (-14.72 = -14.22 + -0.50)

| Location | 5,936,132 – 5,936,249 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -40.71 |
| Consensus MFE | -24.19 |
| Energy contribution | -24.28 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
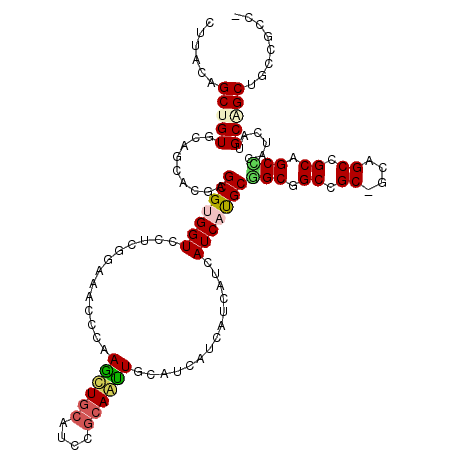
>2L_DroMel_CAF1 5936132 117 + 22407834 CCCCCAGCUGUGCAGCACGAGCGCGGUCCUAGGAAACCCAAGUUGCACCCGCAACUGCAUCAUCAUCAUCAUCAUGCUGCUGCCGCCGCCGCUGCAGCGCAUCAUGCAGCAGCCGCC- ......((...((((((.((..(.(((........)))).((((((....))))))...............)).)))))).)).((.((.((((((........)))))).)).)).- ( -37.90) >DroVir_CAF1 26869 114 + 1 -UUUUAGCUGUGCAGCACGAGCGCGGUCCACGCAAACCGAAGCUGCAUCCGCAGUUGCAUCAUCAUCAUCAUCAUGCGGCCGCUGC---GGCUGCAGCGCAUCAUGCAGCCGCCGCCC -.....((.((((.(((.(((((((.....)))........((((((.(((((((.((..(((.((....)).)))..)).)))))---)).)))))))).)).))).).))).)).. ( -44.80) >DroPse_CAF1 27185 117 + 1 CUUACAGCCGUUCAACACGAGCGUGGUCCUCGGAAACCCAAGCUCCAUCCGCAGUUGCAUCACCAUCAUCAUCACGCGGCGGCUGCAGCAGCCGCUGCCCAUCAUGCCGCUGCACAU- ......(((((.....))).))((((..((.((....)).))..))))..(((((.((((...............((((((((((...)))))))))).....)))).)))))....- ( -39.15) >DroGri_CAF1 24954 114 + 1 -UUCUAGCUGUUCAGCACGAGCGUGGUCCCCGCAAACCAAAGCUGCAUCCGCAAUUGCAUCAUCAUCAUCAUCAUGCGGCGGCCGC---AGCGGCAGCUCAUCAUGCGGCCGCCGCCC -....((((((((.....))))((((...)))).......))))(((........))).................(((((((((((---(((....))......)))))))))))).. ( -41.90) >DroWil_CAF1 32087 117 + 1 UUCUCAGCUGUGCAGCACGAGCGUGGUCCUCGGAAACCGAAAUUGCAUCCCCAAUUGCAUCAUCAUCAUCAUCAUGCGGCAGCCGCUGCGGCAGCGGCUCAUCAUGCAGCUCAUGCC- ......((((.((.(((.(((((((((..((((...))))...((((........))))...........))))))).(.((((((((...))))))))).)).))).)).)).)).- ( -38.20) >DroPer_CAF1 27391 117 + 1 CUUACAGCCGUUCAACACGAGCGUGGUCCUCGGAAACCCAAGCUGCAUCCGCAGUUGCACCACCAUCAUCAUCACGCGGCGGCUGCAGCAGCCGCUGCCCAUCAUGCCGCUGCACAU- ......(((((.....))).))(((((.(..((....)).((((((....))))))).)))))............((((((((.(((((....))))).......))))))))....- ( -42.30) >consensus CUUACAGCUGUGCAGCACGAGCGUGGUCCUCGGAAACCCAAGCUGCAUCCGCAAUUGCAUCAUCAUCAUCAUCAUGCGGCGGCCGC_GCAGCCGCAGCCCAUCAUGCAGCUGCCGCC_ ......(((((.........(((((((.............((((((....))))))..............)))))))(((.((.((....)).)).)))......)))))........ (-24.19 = -24.28 + 0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:57 2006