
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 623,945 – 624,422 |
| Length | 477 |
| Max. P | 0.999904 |

| Location | 623,945 – 624,063 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -30.04 |
| Energy contribution | -30.28 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 623945 118 + 22407834 UCGUA--GCAAACGAAGCAACAGGUUGCUAAGAAACGGGGUUUUUUUUUGUGGGGUUGAGGGGUCUGGAAGCCACUGCUUCUAGUUCCAUUCAACUUCAGCCUAAUGCGUAAAUUUAGUG ((((.--....)))).(((..(((((((.(((((((...)))))))...))(((((((((((..((((((((....))))))))..)).))))))))))))))..)))............ ( -38.50) >DroSec_CAF1 13209 118 + 1 UCGUA--GUAAACGAAGCAACAGGUUGCUAAGAAACGGGGGGUUUAUUUUGGGGGUUAAGGGGGCUGGAAGCCACUGCUUCUAGUUCCAUUCAAUUUCAGCCUAAUGCGUAAAUUUAGUG ((((.--....)))).(((..(((((((((((((((.....)))..))))))........((((((((((((....)))))))))))).........))))))..)))............ ( -34.70) >DroSim_CAF1 12770 118 + 1 UCGUA--GUAAACGAAGCAACAGGUUGCUAUGAAACGGGGGGUUUAUUUUGGGGGUUAAGGGGGCUGGAAGCCACUGCUUCUAGUUCCAUUCAACUUCAGCCUAAUGCGUAAAUUUAGUG (((((--((((.(.........).))))))))).((...((((((((.(((((((((...((((((((((((....))))))))))))....))))....)))))...)))))))).)). ( -36.10) >DroEre_CAF1 16858 114 + 1 UCAUA--CUACCCAAAGCAACAGGUUGCUAAGAAACGGGGGUUU----GUAGGGGUUAAGGGGGCUGGAAGCCCCGGCUUCUAGUUCCAUUCAACUUCAGCCUAAUGCGUAAAUUUAGUG ...((--(((......(((..(((((((((..((((....))))----.)))(((((...((((((((((((....))))))))))))....)))))))))))..))).......))))) ( -37.82) >DroYak_CAF1 15816 114 + 1 UCGUAUCCGACGAACAGCAACAGGUUGCUAAGAAACGUGGUUU------UAGGGGUUAAGGGGGCUGGAAGCCCCUGCUUCUAGUUCCAUUCAACUUCAGCCUAAUGCGUAAAUUUAGUG ((((.....))))...(((..((((((..(((....((((..(------(((((((...((((((.....))))))))))))))..))))....)))))))))..)))............ ( -38.90) >consensus UCGUA__GUAAACGAAGCAACAGGUUGCUAAGAAACGGGGGUUUU_UUUUAGGGGUUAAGGGGGCUGGAAGCCACUGCUUCUAGUUCCAUUCAACUUCAGCCUAAUGCGUAAAUUUAGUG ................(((..(((((.((........))............(..(((...((((((((((((....))))))))))))....)))..))))))..)))............ (-30.04 = -30.28 + 0.24)

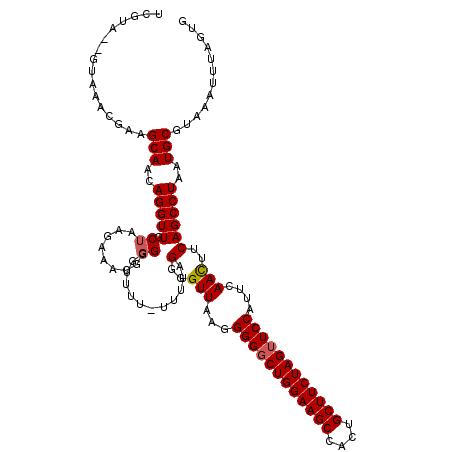
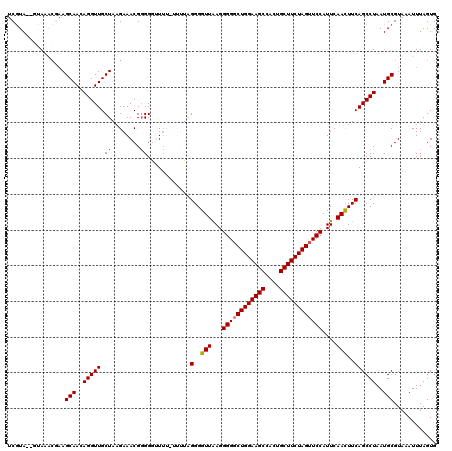
| Location | 623,983 – 624,102 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -40.16 |
| Consensus MFE | -36.90 |
| Energy contribution | -36.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.47 |
| SVM RNA-class probability | 0.999904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 623983 119 + 22407834 UUUUUUUUUGUGGGGUUGAGGGGUCUGGAAGCCACUGCUUCUAGUUCCAUUCAACUUCAGCCUAAUGCGUAAAUUUAGUGAAUGGUCAAUAAGUGGAGACGAAAGAGGAAA-ACCCCGAU .......(((.(((((((((((..((((((((....))))))))..)).)))))).....(((....(((..(((((.(((....))).)))))....)))....)))...-.)))))). ( -37.00) >DroSec_CAF1 13247 119 + 1 GGUUUAUUUUGGGGGUUAAGGGGGCUGGAAGCCACUGCUUCUAGUUCCAUUCAAUUUCAGCCUAAUGCGUAAAUUUAGUGAAUGGUCAAUGAGUGGAGACGAAAGGGGAAA-ACCCCGAU ........((((((......((((((((((((....))))))))))))......((((..(((....(((..(((((.(((....))).)))))....)))..))).))))-.)))))). ( -39.80) >DroSim_CAF1 12808 119 + 1 GGUUUAUUUUGGGGGUUAAGGGGGCUGGAAGCCACUGCUUCUAGUUCCAUUCAACUUCAGCCUAAUGCGUAAAUUUAGUGAAUGGUCAAUAAGUGGAGACGAAAGGGGAAA-ACCCCGAU ...........((((((...((((((((((((....))))))))))))............(((....(((..(((((.(((....))).)))))....)))....)))..)-)))))... ( -39.30) >DroEre_CAF1 16896 116 + 1 GUUU----GUAGGGGUUAAGGGGGCUGGAAGCCCCGGCUUCUAGUUCCAUUCAACUUCAGCCUAAUGCGUAAAUUUAGUGAAUGGUCAAUAAGUGGAGACGAAAGGGGCAAGACCCCGAU ....----...((((((...((((((((((((....))))))))))))...........((((....(((..(((((.(((....))).)))))....)))....))))..))))))... ( -46.00) >DroYak_CAF1 15856 114 + 1 UUU------UAGGGGUUAAGGGGGCUGGAAGCCCCUGCUUCUAGUUCCAUUCAACUUCAGCCUAAUGCGUAAAUUUAGUGAAUGGUCAAUAAGUGCAGACGAGAGAGGAAAACCCCCGAC ..(------..((((((...((((((((((((....))))))))))))............(((....(((..(((((.(((....))).)))))....)))....)))..))))))..). ( -38.70) >consensus GUUUU_UUUUAGGGGUUAAGGGGGCUGGAAGCCACUGCUUCUAGUUCCAUUCAACUUCAGCCUAAUGCGUAAAUUUAGUGAAUGGUCAAUAAGUGGAGACGAAAGGGGAAA_ACCCCGAU ...........(((((....((((((((((((....))))))))))))............(((....(((..(((((.(((....))).)))))....)))....)))....)))))... (-36.90 = -36.90 + -0.00)

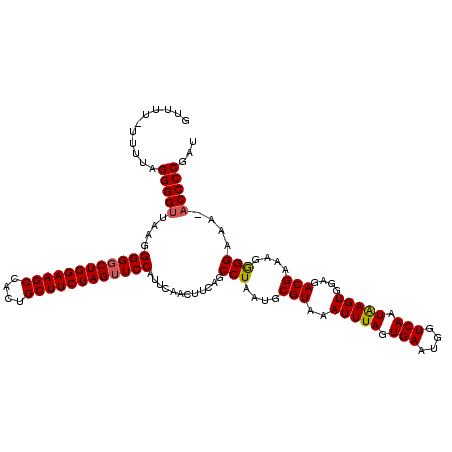
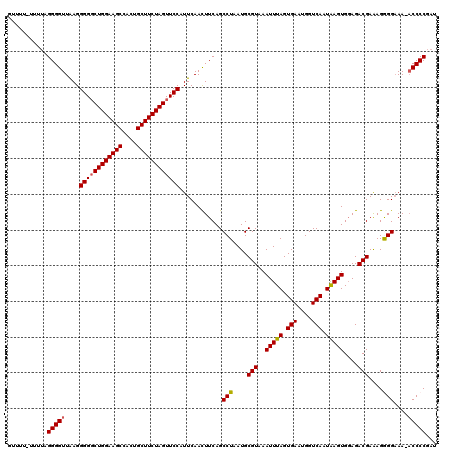
| Location | 623,983 – 624,102 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -24.98 |
| Energy contribution | -25.18 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 623983 119 - 22407834 AUCGGGGU-UUUCCUCUUUCGUCUCCACUUAUUGACCAUUCACUAAAUUUACGCAUUAGGCUGAAGUUGAAUGGAACUAGAAGCAGUGGCUUCCAGACCCCUCAACCCCACAAAAAAAAA ...(((((-(.........................(((((((.....((((.((.....))))))..)))))))..((.(((((....))))).)).......))))))........... ( -27.70) >DroSec_CAF1 13247 119 - 1 AUCGGGGU-UUUCCCCUUUCGUCUCCACUCAUUGACCAUUCACUAAAUUUACGCAUUAGGCUGAAAUUGAAUGGAACUAGAAGCAGUGGCUUCCAGCCCCCUUAACCCCCAAAAUAAACC ...(((((-(.........................(((((((.....((((.((.....))))))..)))))))..((.(((((....))))).)).......))))))........... ( -24.90) >DroSim_CAF1 12808 119 - 1 AUCGGGGU-UUUCCCCUUUCGUCUCCACUUAUUGACCAUUCACUAAAUUUACGCAUUAGGCUGAAGUUGAAUGGAACUAGAAGCAGUGGCUUCCAGCCCCCUUAACCCCCAAAAUAAACC ...(((((-(.........................(((((((.....((((.((.....))))))..)))))))..((.(((((....))))).)).......))))))........... ( -26.00) >DroEre_CAF1 16896 116 - 1 AUCGGGGUCUUGCCCCUUUCGUCUCCACUUAUUGACCAUUCACUAAAUUUACGCAUUAGGCUGAAGUUGAAUGGAACUAGAAGCCGGGGCUUCCAGCCCCCUUAACCCCUAC----AAAC ...(((((...(((((....(((..........)))......................((((..((((......))))...))))))))).....)))))............----.... ( -30.20) >DroYak_CAF1 15856 114 - 1 GUCGGGGGUUUUCCUCUCUCGUCUGCACUUAUUGACCAUUCACUAAAUUUACGCAUUAGGCUGAAGUUGAAUGGAACUAGAAGCAGGGGCUUCCAGCCCCCUUAACCCCUA------AAA ...(((((((.............(((.(.....).(((((((.....((((.((.....))))))..)))))))........)))(((((.....)))))...))))))).------... ( -32.60) >consensus AUCGGGGU_UUUCCCCUUUCGUCUCCACUUAUUGACCAUUCACUAAAUUUACGCAUUAGGCUGAAGUUGAAUGGAACUAGAAGCAGUGGCUUCCAGCCCCCUUAACCCCCAAAA_AAAAC ...(((((...........................(((((((.....((((.((.....))))))..)))))))..((.(((((....))))).))........)))))........... (-24.98 = -25.18 + 0.20)

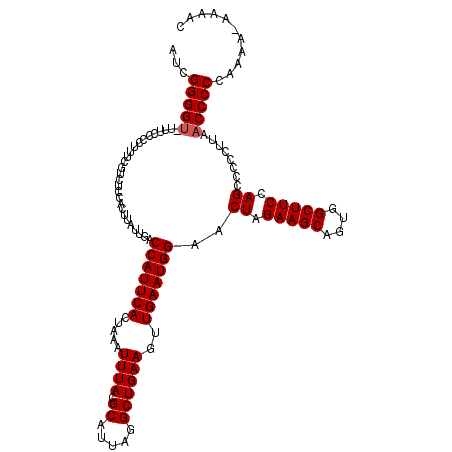
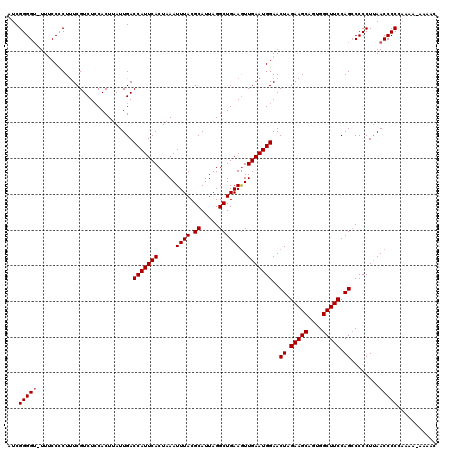
| Location | 624,063 – 624,182 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.07 |
| Mean single sequence MFE | -40.26 |
| Consensus MFE | -36.73 |
| Energy contribution | -36.69 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 624063 119 + 22407834 AAUGGUCAAUAAGUGGAGACGAAAGAGGAAA-ACCCCGAUGUUGGUGAUGUGGCCAUCGCAGUGGCAAUGUGUUGCUGUGGUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUC .....((((((..(((...(....).((...-.))))).))))))..((((.((((((((((..((.....))..))))))))(((((((....))))))).)).))))........... ( -38.80) >DroSec_CAF1 13327 119 + 1 AAUGGUCAAUGAGUGGAGACGAAAGGGGAAA-ACCCCGAUGUUGGUGAUGUGGCCGUCGCAGUGGCACUGUGUUGCUGUGGUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUC ....((((.(.(((...(((....((((...-.))))......(((......))))))((((..((((((..((.....))..))).)))..)))).))).).))))............. ( -42.10) >DroSim_CAF1 12888 119 + 1 AAUGGUCAAUAAGUGGAGACGAAAGGGGAAA-ACCCCGAUGUUGGUGAUGUGGCCGUCGCAGUGGCACUGUGUUGCUGUGGUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUC ....((((...(((...(((....((((...-.))))......(((......))))))((((..((((((..((.....))..))).)))..)))).)))...))))............. ( -42.20) >DroEre_CAF1 16972 120 + 1 AAUGGUCAAUAAGUGGAGACGAAAGGGGCAAGACCCCGAUGUUGGUGAUGUGGCCGUAGCAGUGGCAAUGUGUUGCUGUGGUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUC .(((((((....((.(.((((...((((.....))))..))))..).)).))))))).((((..(((((....(((......))))))))..)))).(((((.(((....))).))))). ( -40.70) >DroYak_CAF1 15930 120 + 1 AAUGGUCAAUAAGUGCAGACGAGAGAGGAAAACCCCCGACGUUGGUGAUGUGGCCGUCACAGUGGCAAUGUGUUACUGUGGUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUC .(((((((.....(((.((((...(.((......)))..)))).)))...)))))))(((((((((.....))))))))).((((((....))))))(((((.(((....))).))))). ( -37.50) >consensus AAUGGUCAAUAAGUGGAGACGAAAGGGGAAA_ACCCCGAUGUUGGUGAUGUGGCCGUCGCAGUGGCAAUGUGUUGCUGUGGUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUC .(((((((.........((((...((((.....))))..)))).......)))))))(((((((((.....))))))))).((((((....))))))(((((.(((....))).))))). (-36.73 = -36.69 + -0.04)

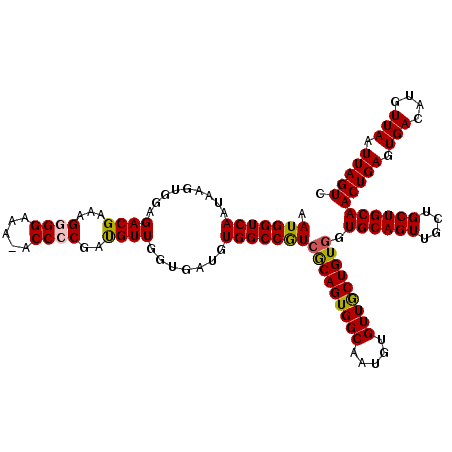
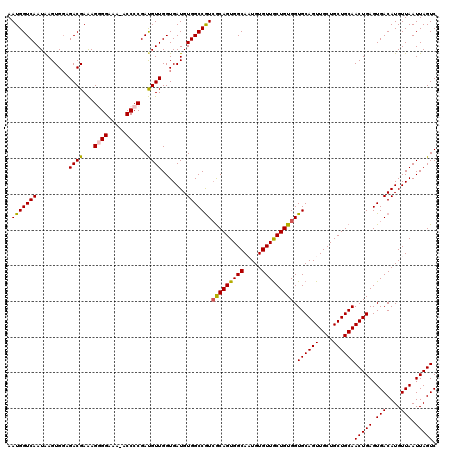
| Location | 624,063 – 624,182 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.07 |
| Mean single sequence MFE | -26.31 |
| Consensus MFE | -24.16 |
| Energy contribution | -23.48 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 624063 119 - 22407834 GACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCACCACAGCAACACAUUGCCACUGCGAUGGCCACAUCACCAACAUCGGGGU-UUUCCUCUUUCGUCUCCACUUAUUGACCAUU (((.........(((....(((((((....)))))))....)))((((....)))).(((.(((((.............))))).)))-...........)))................. ( -23.62) >DroSec_CAF1 13327 119 - 1 GACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCACCACAGCAACACAGUGCCACUGCGACGGCCACAUCACCAACAUCGGGGU-UUUCCCCUUUCGUCUCCACUCAUUGACCAUU (((........((((......(((((((..(((..(((......)))......)))..)))))))((........)).)))).((((.-...))))....)))................. ( -27.30) >DroSim_CAF1 12888 119 - 1 GACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCACCACAGCAACACAGUGCCACUGCGACGGCCACAUCACCAACAUCGGGGU-UUUCCCCUUUCGUCUCCACUUAUUGACCAUU (((........((((......(((((((..(((..(((......)))......)))..)))))))((........)).)))).((((.-...))))....)))................. ( -27.30) >DroEre_CAF1 16972 120 - 1 GACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCACCACAGCAACACAUUGCCACUGCUACGGCCACAUCACCAACAUCGGGGUCUUGCCCCUUUCGUCUCCACUUAUUGACCAUU (((.........(((....(((((((....)))))))....)))((((....))))....((....))...............((((.....))))....)))................. ( -25.30) >DroYak_CAF1 15930 120 - 1 GACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCACCACAGUAACACAUUGCCACUGUGACGGCCACAUCACCAACGUCGGGGGUUUUCCUCUCUCGUCUGCACUUAUUGACCAUU (((..........)))...(((((((....)))))))...((((((..(.....)..))))))..((.((......(((((..(((((....)))))..))).)).......)).))... ( -28.02) >consensus GACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCACCACAGCAACACAUUGCCACUGCGACGGCCACAUCACCAACAUCGGGGU_UUUCCCCUUUCGUCUCCACUUAUUGACCAUU (((........((((......(((((((..(((..(((......)))......)))..)))))))((........)).)))).((((.....))))....)))................. (-24.16 = -23.48 + -0.68)

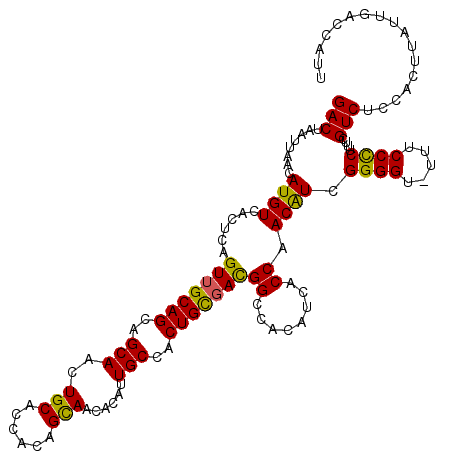
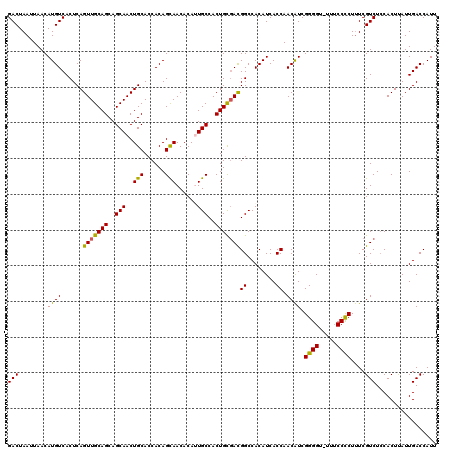
| Location | 624,102 – 624,222 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -54.16 |
| Consensus MFE | -52.66 |
| Energy contribution | -52.34 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.05 |
| Mean z-score | -5.28 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 624102 120 + 22407834 GUUGGUGAUGUGGCCAUCGCAGUGGCAAUGUGUUGCUGUGGUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUCGCUUACAUGACAGCAGUUUGCUGCCAAGGCAUUGCCAAAA .((((..((((....(((((((..((.....))..)))))))(((((.(((((((((..((((((((..........))))))))..)).)))))))..)))))....))))..)))).. ( -55.50) >DroSec_CAF1 13366 120 + 1 GUUGGUGAUGUGGCCGUCGCAGUGGCACUGUGUUGCUGUGGUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUCGCUUACAUGACAGCAGUUUGCUGCCAAGGCAUUGCCAAAA .((((..((((......(((((..((.....))..)))))(.(((((.(((((((((..((((((((..........))))))))..)).)))))))..))))))...))))..)))).. ( -53.90) >DroSim_CAF1 12927 120 + 1 GUUGGUGAUGUGGCCGUCGCAGUGGCACUGUGUUGCUGUGGUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUCGCUUACAUGACAGCAGUUUGCUGCCAAGGCAUUGCCAAAA .((((..((((......(((((..((.....))..)))))(.(((((.(((((((((..((((((((..........))))))))..)).)))))))..))))))...))))..)))).. ( -53.90) >DroEre_CAF1 17012 120 + 1 GUUGGUGAUGUGGCCGUAGCAGUGGCAAUGUGUUGCUGUGGUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUCGCUUACAUGACAGCAGUUUGCUGCCAAGGCAUUGCCAAAA .((((..((((.(((...((((..((.....))..)))))))(((((.(((((((((..((((((((..........))))))))..)).)))))))..)))))....))))..)))).. ( -53.40) >DroYak_CAF1 15970 120 + 1 GUUGGUGAUGUGGCCGUCACAGUGGCAAUGUGUUACUGUGGUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUCGCUUACAUGACAGCAGUUUGCUGCCAAGGCAUUGCCAAAA .((((..((((......(((((((((.....)))))))))(.(((((.(((((((((..((((((((..........))))))))..)).)))))))..))))))...))))..)))).. ( -54.10) >consensus GUUGGUGAUGUGGCCGUCGCAGUGGCAAUGUGUUGCUGUGGUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUCGCUUACAUGACAGCAGUUUGCUGCCAAGGCAUUGCCAAAA .((((..((((.(((...((((((((.....)))))))))))(((((.(((((((((..((((((((..........))))))))..)).)))))))..)))))....))))..)))).. (-52.66 = -52.34 + -0.32)

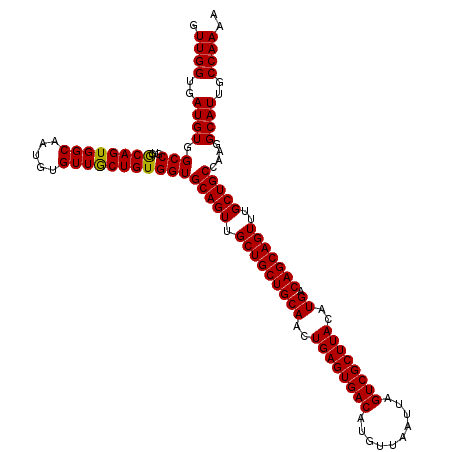
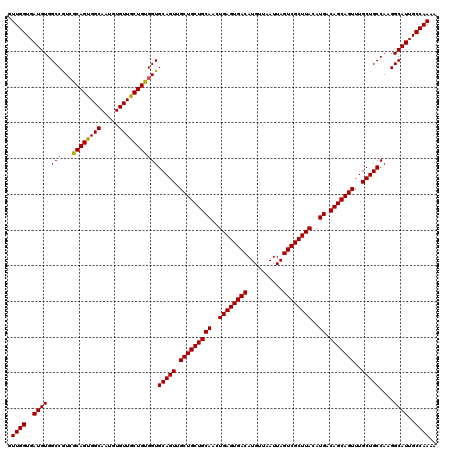
| Location | 624,102 – 624,222 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -35.26 |
| Energy contribution | -35.54 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.12 |
| SVM RNA-class probability | 0.999806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 624102 120 - 22407834 UUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGCGACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCACCACAGCAACACAUUGCCACUGCGAUGGCCACAUCACCAAC ...((((((((...(((((((.....))))))).....(((((..........)))...(((((((....))))))).......))....))))))))....((((....))))...... ( -40.00) >DroSec_CAF1 13366 120 - 1 UUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGCGACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCACCACAGCAACACAGUGCCACUGCGACGGCCACAUCACCAAC ...((((..(((..(((((......((((((..(((.((.(((..........))).)).((((((....)))))))))..))))))......)))))..)))...)))).......... ( -36.30) >DroSim_CAF1 12927 120 - 1 UUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGCGACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCACCACAGCAACACAGUGCCACUGCGACGGCCACAUCACCAAC ...((((..(((..(((((......((((((..(((.((.(((..........))).)).((((((....)))))))))..))))))......)))))..)))...)))).......... ( -36.30) >DroEre_CAF1 17012 120 - 1 UUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGCGACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCACCACAGCAACACAUUGCCACUGCUACGGCCACAUCACCAAC ...((((((((...(((((((.....))))))).....(((((..........)))...(((((((....))))))).......))....))))))))..((....))............ ( -38.50) >DroYak_CAF1 15970 120 - 1 UUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGCGACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCACCACAGUAACACAUUGCCACUGUGACGGCCACAUCACCAAC ...((((((((...(((((((.....)))))))(((.((.(((..........))).))(((((((....)))))))....)))......))))))))..((((.(....).)))).... ( -37.40) >consensus UUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGCGACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCACCACAGCAACACAUUGCCACUGCGACGGCCACAUCACCAAC ...((((..(((..((((((.....((((((..(((.((.(((..........))).)).((((((....)))))))))..)))))).....))))))..)))...)))).......... (-35.26 = -35.54 + 0.28)

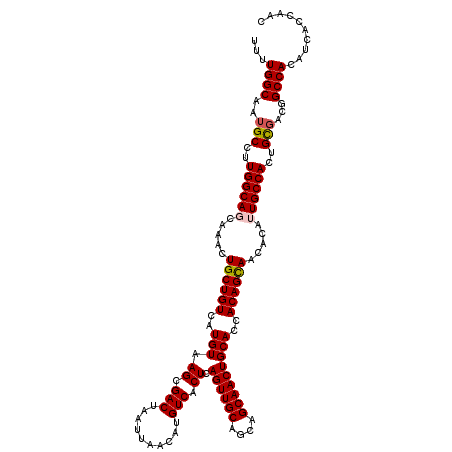
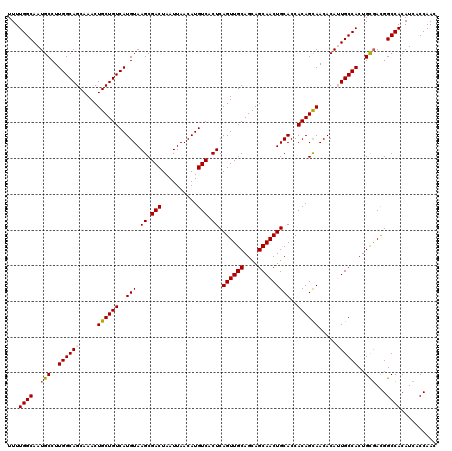
| Location | 624,142 – 624,262 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -46.18 |
| Consensus MFE | -44.66 |
| Energy contribution | -44.86 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.18 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.999271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 624142 120 + 22407834 GUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUCGCUUACAUGACAGCAGUUUGCUGCCAAGGCAUUGCCAAAAUGCUAAUUUCCACAUUGGGCAACCGCUGAGACGCGGAAAA (.(((((.(((((((((..((((((((..........))))))))..)).)))))))..))))))..((((((.....))))))..(((((.....((....))((......))))))). ( -46.50) >DroSec_CAF1 13406 119 + 1 GUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUCGCUUACAUGACAGCAGUUUGCUGCCAAGGCAUUGCCAAAAUGCUAAUUUCCACAUUG-GCAACCGCUGAGACGCGGAAAA (.(((((.(((((((((..((((((((..........))))))))..)).)))))))..))))))..(((...)))....(((((((......))))-))).((((......)))).... ( -44.90) >DroSim_CAF1 12967 120 + 1 GUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUCGCUUACAUGACAGCAGUUUGCUGCCAAGGCAUUGCCAAAAUGCUAAUUUCCACAUUGGGCAACCGCUGAGACGCGGAAAA (.(((((.(((((((((..((((((((..........))))))))..)).)))))))..))))))..((((((.....))))))..(((((.....((....))((......))))))). ( -46.50) >DroEre_CAF1 17052 120 + 1 GUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUCGCUUACAUGACAGCAGUUUGCUGCCAAGGCAUUGCCAAAAUGCUAAUUUCCACAUUGGGCAACCGCUGAGACGCGGAAAA (.(((((.(((((((((..((((((((..........))))))))..)).)))))))..))))))..((((((.....))))))..(((((.....((....))((......))))))). ( -46.50) >DroYak_CAF1 16010 120 + 1 GUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUCGCUUACAUGACAGCAGUUUGCUGCCAAGGCAUUGCCAAAAUGCUAAUUUCCACAUUGGGCAACCGCUGAGACGCGGAAAA (.(((((.(((((((((..((((((((..........))))))))..)).)))))))..))))))..((((((.....))))))..(((((.....((....))((......))))))). ( -46.50) >consensus GUGCAGUUGCUGCUGCAACUGAGUGACAUGUUAAUUAGUCGCUUACAUGACAGCAGUUUGCUGCCAAGGCAUUGCCAAAAUGCUAAUUUCCACAUUGGGCAACCGCUGAGACGCGGAAAA (.(((((.(((((((((..((((((((..........))))))))..)).)))))))..))))))..((((((.....))))))..(((((.....((....))((......))))))). (-44.66 = -44.86 + 0.20)

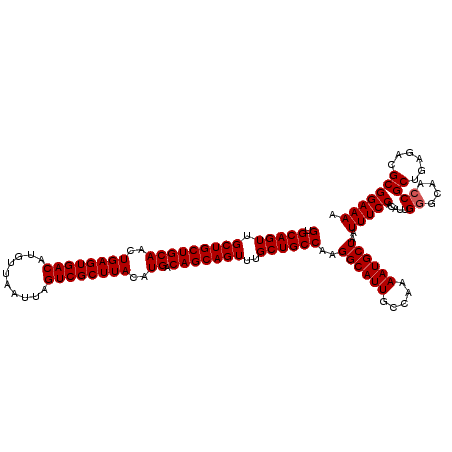
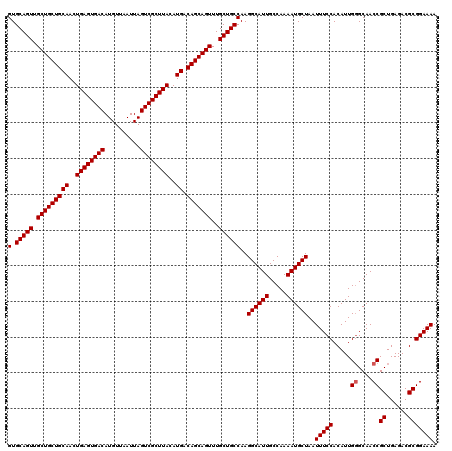
| Location | 624,142 – 624,262 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -39.36 |
| Consensus MFE | -37.64 |
| Energy contribution | -37.84 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 624142 120 - 22407834 UUUUCCGCGUCUCAGCGGUUGCCCAAUGUGGAAAUUAGCAUUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGCGACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCAC ....((((......))))(((((.(((((........)))))..)))))(((..(((((((.....))))))).)))((.(((..........))).))(((((((....)))))))... ( -39.00) >DroSec_CAF1 13406 119 - 1 UUUUCCGCGUCUCAGCGGUUGC-CAAUGUGGAAAUUAGCAUUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGCGACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCAC ....((((......))))((((-(((.(((........))).)))))))(((..(((((((.....))))))).)))((.(((..........))).))(((((((....)))))))... ( -40.80) >DroSim_CAF1 12967 120 - 1 UUUUCCGCGUCUCAGCGGUUGCCCAAUGUGGAAAUUAGCAUUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGCGACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCAC ....((((......))))(((((.(((((........)))))..)))))(((..(((((((.....))))))).)))((.(((..........))).))(((((((....)))))))... ( -39.00) >DroEre_CAF1 17052 120 - 1 UUUUCCGCGUCUCAGCGGUUGCCCAAUGUGGAAAUUAGCAUUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGCGACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCAC ....((((......))))(((((.(((((........)))))..)))))(((..(((((((.....))))))).)))((.(((..........))).))(((((((....)))))))... ( -39.00) >DroYak_CAF1 16010 120 - 1 UUUUCCGCGUCUCAGCGGUUGCCCAAUGUGGAAAUUAGCAUUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGCGACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCAC ....((((......))))(((((.(((((........)))))..)))))(((..(((((((.....))))))).)))((.(((..........))).))(((((((....)))))))... ( -39.00) >consensus UUUUCCGCGUCUCAGCGGUUGCCCAAUGUGGAAAUUAGCAUUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGCGACUAAUUAACAUGUCACUCAGUUGCAGCAGCAACUGCAC ....((((......))))(((((.(((((........)))))..)))))(((..(((((((.....))))))).)))((.(((..........))).))(((((((....)))))))... (-37.64 = -37.84 + 0.20)

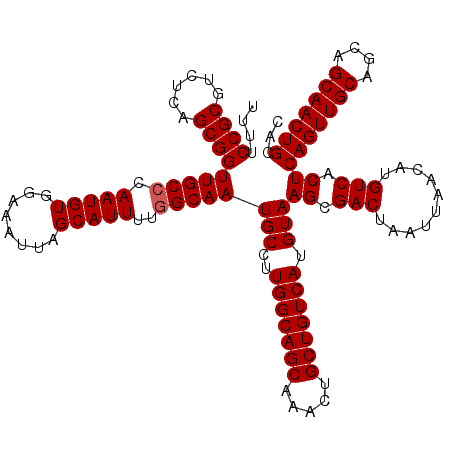
| Location | 624,182 – 624,302 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -35.74 |
| Consensus MFE | -32.02 |
| Energy contribution | -32.26 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 624182 120 - 22407834 CACAACUCCGCGAGCCCGGCUCCUUUUCCAUUUUCCGAGCUUUUCCGCGUCUCAGCGGUUGCCCAAUGUGGAAAUUAGCAUUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGC .........((..(((..(((...(((((((.....(.((....((((......))))..)))....)))))))..))).....)))..(((..(((((((.....))))))).))).)) ( -36.70) >DroSec_CAF1 13446 119 - 1 CACAACUCCGCGAACCCGGCUCUUUUCCCAUUUUCCGCGCUUUUCCGCGUCUCAGCGGUUGC-CAAUGUGGAAAUUAGCAUUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGC .........((.((....(((..(((((((((....(.((....((((......))))..))-))))).)))))..)))...)).))..(((..(((((((.....))))))).)))... ( -35.20) >DroSim_CAF1 13007 120 - 1 CACAACUACGCGAACCAGGCUCUUUUUCCAUUUUCCGCGCUUUUCCGCGUCUCAGCGGUUGCCCAAUGUGGAAAUUAGCAUUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGC .........((.((....(((...(((((((.....(.((....((((......))))..)))....)))))))..)))...)).))..(((..(((((((.....))))))).)))... ( -34.70) >DroEre_CAF1 17092 120 - 1 CACAACUCUGCGAACCCGACUCCUUUUCCAUAUUCCGCGCUUUUCCGCGUCUCAGCGGUUGCCCAAUGUGGAAAUUAGCAUUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGC ........(((.((...(.((...((((((((((..(((.....((((......)))).)))..))))))))))..)))...)).))).(((..(((((((.....))))))).)))... ( -34.20) >DroYak_CAF1 16050 120 - 1 CACAACUCCGCGAACCCGGCUCGUUUUCCAUAUUCCGCGCUUUUCCGCGUCUCAGCGGUUGCCCAAUGUGGAAAUUAGCAUUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGC .........((.((....(((...((((((((((..(((.....((((......)))).)))..))))))))))..)))...)).))..(((..(((((((.....))))))).)))... ( -37.90) >consensus CACAACUCCGCGAACCCGGCUCCUUUUCCAUUUUCCGCGCUUUUCCGCGUCUCAGCGGUUGCCCAAUGUGGAAAUUAGCAUUUUGGCAAUGCCUUGGCAGCAAACUGCUGUCAUGUAAGC .........((.((....(((...(((((((.......((....((((......))))..)).....)))))))..)))...)).))..(((..(((((((.....))))))).)))... (-32.02 = -32.26 + 0.24)

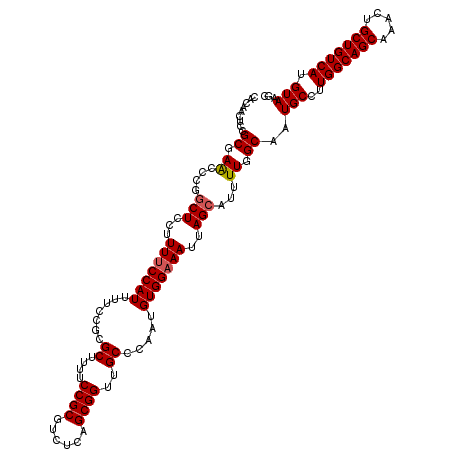
| Location | 624,302 – 624,422 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -24.80 |
| Energy contribution | -24.52 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 624302 120 + 22407834 CAAUUUAUAAUUGAGCAAUUUGCCAGCGACUGAUUUAUUUGUGGAUCUCCAAAUGGGAUUAAAACGGCUUGUGUCUCUCCAAUGCAAUAUGAAUCGCUGGGAAAACAAGAUCAAAUGUAC .....((((.((((....(((.(((((((.(.((.......((((((.((....)))).......(((....)))..)))).......)).).))))))).)))......)))).)))). ( -26.74) >DroSec_CAF1 13565 120 + 1 CAAUUUAUAAUUGAGCAAUUUUCCAGCGACUGAUUUAUUUGUGGAUCUCCAAAUGGGAUUAACACGGCUUGUGUCUCUCCAAUGCAAUAUGAAUCGCUGGGAAAACAAGAUCAAAUGUAC .....((((.((((....(((((((((((....(((((((((((....)).....(((...((((.....))))...)))...)))).))))))))))))))))......)))).)))). ( -29.70) >DroSim_CAF1 13127 120 + 1 CAAUUUAUAAUUGAGCAAUUUGCCAGCGACUGAUUUAUUUGUGGAUCUCCAAAUGGGAUUAAAACGGCUUGUGUCUCUCCAAUGCAAUAUGAAUCGCUGGGAAAACAAGAGCAAAUGUAC .....((((.(((..(..(((.(((((((.(.((.......((((((.((....)))).......(((....)))..)))).......)).).))))))).)))....)..))).)))). ( -25.74) >DroEre_CAF1 17212 118 + 1 CAAUUUAUAAUUGAGCAAUUUGCCAGCGACUGAUUUAUUUGUGGAUCCCCACAUGGGAUUAAAACGGUCUGUGUC--UCCAAUGCAAUAUGAAUCGCUGGGAAAACAAGAUCAAAUGUAC .....((((.((((....(((.((((((((((.((((..(((((....)))))......)))).)))))(((((.--....))))).........))))).)))......)))).)))). ( -28.50) >DroYak_CAF1 16170 120 + 1 CAAUUUAUAAUUGAGCAAUUUGCCAGCGACUGAUUUAUUUGUGGAUCCCCAAAUGGGAUUAAAACGGUCUGUGUCUCUCCAAUGCAACAUGAAUCGCUGGGAAAACAAGAUCAAAUGUAC .....((((.((((....(((.(((((((.........((((.((((((.....))))))...))))((.((((..(......)..)))))).))))))).)))......)))).)))). ( -28.40) >consensus CAAUUUAUAAUUGAGCAAUUUGCCAGCGACUGAUUUAUUUGUGGAUCUCCAAAUGGGAUUAAAACGGCUUGUGUCUCUCCAAUGCAAUAUGAAUCGCUGGGAAAACAAGAUCAAAUGUAC .....((((.((((....(((.(((((((..........(((.((((((.....))))))...)))((...((......))..))........))))))).)))......)))).)))). (-24.80 = -24.52 + -0.28)

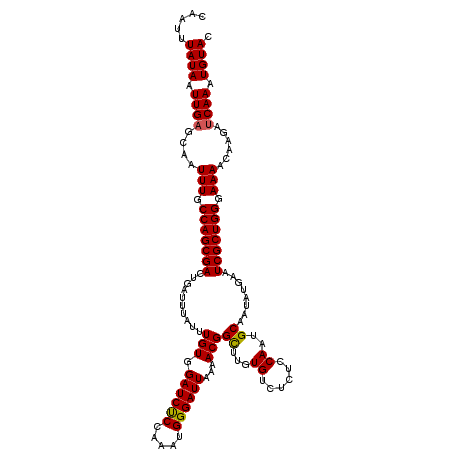
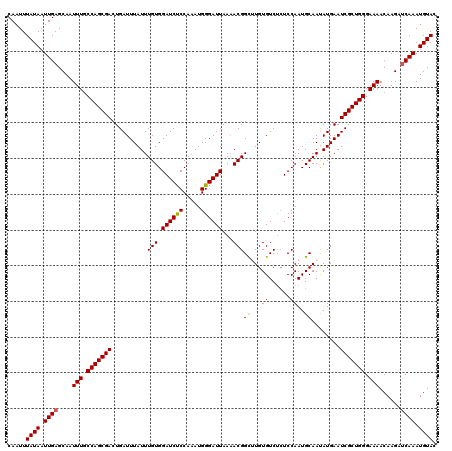
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:24 2006