| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,909,990 – 5,910,110 |
| Length | 120 |
| Max. P | 0.753554 |

| Location | 5,909,990 – 5,910,110 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.94 |
| Mean single sequence MFE | -45.21 |
| Consensus MFE | -28.12 |
| Energy contribution | -29.52 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
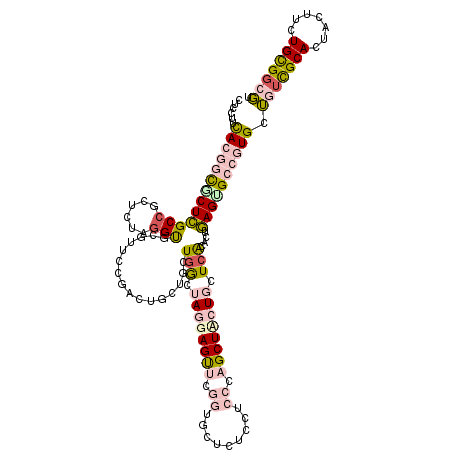
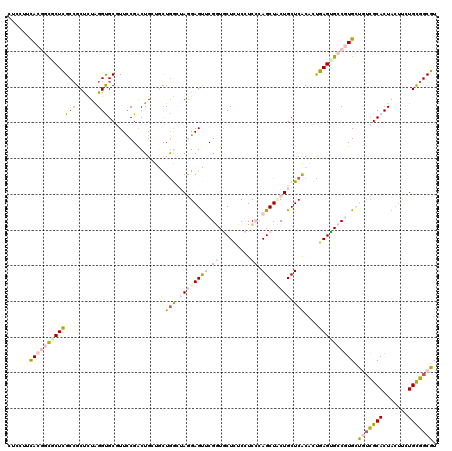
>2L_DroMel_CAF1 5909990 120 - 22407834 GUCCUUUACGGCGCUCGCCGCUCUAGGCGCGUUCCGACUGCUAUUGGCUAGGAGUACGGUGCUCUCCUCCCAGCUCCUGCUCACACUGAGUGCCGUGCUGUCGCACUAUUUCUGCGGCGU .((((...(((.((.((((......)))).)).)))...((.....)).))))((((((..(((.......(((....)))......)))..)))))).((((((.......)))))).. ( -47.62) >DroPse_CAF1 207394 120 - 1 CUCCUACACUCUCCUGGCCGCCUUGGGUGCCUUCCGACUGCUGCUGAAAAAGAGUUCGAUACUGUCAUCAGAGCUACUGCUCGCCGCCAGUGCCAUGGGCUUGCACUAUUUCUGUGGCCU ...............((((((....(((((......(((((.(((((...(.(((.....))).)..)))((((....)))))).).))))(((...)))..)))))......)))))). ( -34.00) >DroSec_CAF1 100427 120 - 1 GUCCUUCACGGCGCUCGCAGCUCUAGGCGCGUUUCGACUGGUGCUGGCUAGGAGUACGGUGCUGUCCUCCCAGCUCCUGCUCACACUGAGCGCCGUGCUGUCGCACUACAUUUGCGGCGU ......((((((((((.((((.((((.((.....)).)))).)))).....(((((.((.((((......)))).))))))).....))))))))))..((((((.......)))))).. ( -55.50) >DroEre_CAF1 125150 120 - 1 GUCAUUCACGGCGCUCGCCGCUCUAGGUGCGUUCCGGCUGAUCCUAGCUAGGAGUUCGGUGCUCUCCUCCCAGCUUCUGCUCACACUGAGUGCCGUGCUGGCGCACUACUCCUGCGGCGU (((......)))...((((((....(((((((((((((((....))))).)))((.(((..(((.......(((....)))......)))..))).))..)))))))......)))))). ( -47.82) >DroYak_CAF1 126229 120 - 1 CUCCUUCACGGCACUCGCCGCUCUAGGUGCGUUCCGGCUGAUGCUGGCUAGGAGUUCGGUGCUCUCCUCCCAGCUUCUGCUCACACUGAGUGCCGUGCUGUCGCACUACUUCUGCGGCGU ......(((((((((((((......)))..((...(((.((.(((((..(((((.........))))).))))).)).)))...)).))))))))))..((((((.......)))))).. ( -50.40) >DroPer_CAF1 207339 120 - 1 CUCCUACACUCUCCUGGCCGCCUUGGGUGCCUUCCGACUGCUGCUGAAAAAGAGCUCGAUACUGUCAUCAGAGCUACUGCUCGCCGCCAGUGCCAUGGGCUUGCACUAUUCCUGUGGCCU ...............((((((....(((((......(((((.((.((...((((((((((......))).))))).))..)))).).))))(((...)))..)))))......)))))). ( -35.90) >consensus CUCCUUCACGGCGCUCGCCGCUCUAGGUGCGUUCCGACUGCUGCUGGCUAGGAGUUCGGUGCUCUCCUCCCAGCUACUGCUCACACUGAGUGCCGUGCUGUCGCACUACUUCUGCGGCGU ......(((((((((((((......)))................(((.((((((((.((.........)).)))))))).)))....)))))))))).(((((((.......))))))). (-28.12 = -29.52 + 1.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:46 2006