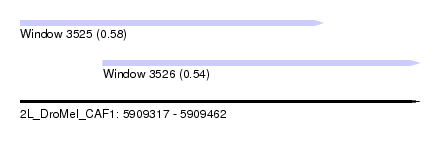
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,909,317 – 5,909,462 |
| Length | 145 |
| Max. P | 0.578342 |

| Location | 5,909,317 – 5,909,427 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 92.12 |
| Mean single sequence MFE | -42.20 |
| Consensus MFE | -36.78 |
| Energy contribution | -37.52 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
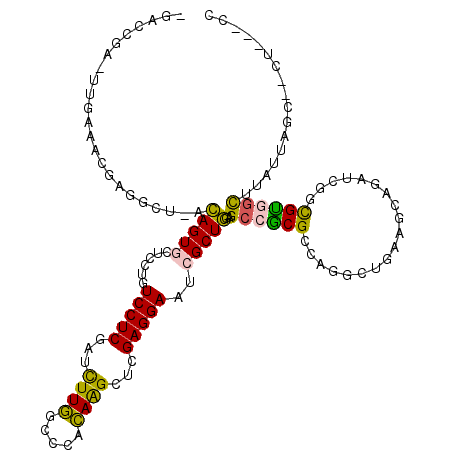
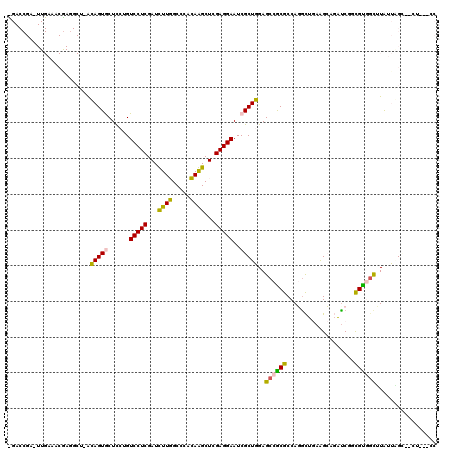
>2L_DroMel_CAF1 5909317 110 + 22407834 GGGUUUCAUUGUGGCCUCGCUGUUCUCACUUGACCGAGUUGAAACAAGGCUAACAGUGCUCCUGUCCUCGAUUUUGGCCCACAAGCUCGAGGAAUCGCUGGAGCCGCGCC .(((...((((((((((...((((.((((((....))).)))))))))))).)))))(((((..(((((((.((((.....)))).)))))))......)))))...))) ( -38.50) >DroSec_CAF1 99754 110 + 1 GGGUUUCACUGUGGCCUCGCUGCUCUCACUUGACCGAUUUGAAACGAUGCUCACAGUGCUCCUGUCCUCGAUUUUGGCCCACAAGCUCGAGGAAUCGCUGGAGCCGCGCC .(((...((((((((.(((...(..((........))...)...))).)).))))))(((((..(((((((.((((.....)))).)))))))......)))))...))) ( -36.50) >DroEre_CAF1 124477 110 + 1 GGGUUUCACUGUGGCCUCGCUGUUCUGACUCGACCGAGUUGACACGAGGCUAGCAGUGCUUCUGUCCUCGAUUUUGGCCCACAAGCUGGAGGAAUCGCUGGAGCCGCGCC .((((((((((((((((((.((((..(((((....))))))))))))))))).))))((.....(((((.(.((((.....)))).).)))))...)).))))))..... ( -46.20) >DroYak_CAF1 125556 110 + 1 GGGUUUCACUGUGGCCUCGCUGCUCUGACUCGACCGAUUUGAAGCGAGGCUUACAGUGCUUCUGUCCUCGAUCUUGGCCCACAGGCUCGAGGAAUCGCUGGAGCCGCGCC .((((((((((((((((((((..((.((.((....)))).)))))))))).))))))((.....(((((((.((((.....)))).)))))))...)).))))))..... ( -47.60) >consensus GGGUUUCACUGUGGCCUCGCUGCUCUCACUCGACCGAGUUGAAACGAGGCUAACAGUGCUCCUGUCCUCGAUUUUGGCCCACAAGCUCGAGGAAUCGCUGGAGCCGCGCC .((((((((((((((((((....((.(((((....)))))))..))))))).)))))((.....(((((((.((((.....)))).)))))))...)).))))))..... (-36.78 = -37.52 + 0.75)

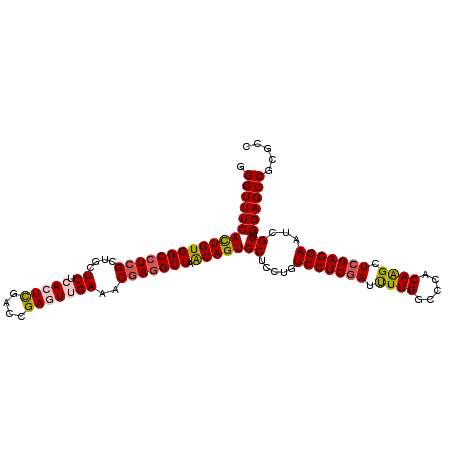
| Location | 5,909,347 – 5,909,462 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.69 |
| Mean single sequence MFE | -41.38 |
| Consensus MFE | -20.73 |
| Energy contribution | -19.38 |
| Covariance contribution | -1.36 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5909347 115 + 22407834 UGACCGAGUUGAAACAAGGCUAACAGUGCUCCUGUCCUCGAUUUUGGCCCACAAGCUCGAGGAAUCGCUGGAGCCGCGCCAGGCUGAAGCAGACCUGCGCAUCUUAUUGGC--CU---GC ................(((((((..(.(((((..(((((((.((((.....)))).)))))))......))))))((((.((((((...))).)))))))......)))))--))---.. ( -45.10) >DroPse_CAF1 206767 93 + 1 ----------------------GGAGUACUCCAGUCCUCGAUCUUUGCCCAAAGGCUAGAGGAAUCGCUUGAGCCACGCCAUGUCCAGGA--GUCCCUGUGGCGUGUGCGUCCCU---CA ----------------------(((....)))..(((((..(((((....)))))...)))))......((((..((((((((((((((.--...)))).)))))).))))..))---)) ( -36.50) >DroSec_CAF1 99784 115 + 1 UGACCGAUUUGAAACGAUGCUCACAGUGCUCCUGUCCUCGAUUUUGGCCCACAAGCUCGAGGAAUCGCUGGAGCCGCGCCAGACUGAAGCUGAUCGGCGCAGCUUAUUGGC--CU---GC ...(((((..(.......((((.(((((......(((((((.((((.....)))).)))))))..))))))))).(((((.(((.......).)))))))..)..))))).--..---.. ( -40.50) >DroEre_CAF1 124507 115 + 1 CGACCGAGUUGACACGAGGCUAGCAGUGCUUCUGUCCUCGAUUUUGGCCCACAAGCUGGAGGAAUCGCUGGAGCCGCGCCAGGCUGAAGCAGAUCGGCGUGGCAUAUUAGG--CU---CC (((.((((..((((.(((((.......))))))))))))).(((..((......))..)))...)))..((((((..((((.(((((......))))).))))......))--))---)) ( -46.30) >DroYak_CAF1 125586 115 + 1 CGACCGAUUUGAAGCGAGGCUUACAGUGCUUCUGUCCUCGAUCUUGGCCCACAGGCUCGAGGAAUCGCUGGAGCCGCGCCAGGCUGAGGCAGAUCGUCGUGGUUUAUUAGG--CU---CC ............((((((((.......)))))..(((((((.((((.....)))).)))))))...)))((((((..((((.(.(((......))).).))))......))--))---)) ( -43.40) >DroPer_CAF1 206709 96 + 1 ----------------------GGAGUACUCCAGUCCUCGAUCUUUGCCCAAAGGCUAGAGGAAUCGCUUGAGCCACGCCAUGUCCAGGA--GUCCCUGUGGCGUGUGCGUCCCUCUCCA ----------------------((((..(((.(((((((..(((((....)))))...))))....))).)))..((((((((((((((.--...)))).)))))).))))....)))). ( -36.50) >consensus _GACCGA_UUGAAACGAGGCU_ACAGUGCUCCUGUCCUCGAUCUUGGCCCACAAGCUCGAGGAAUCGCUGGAGCCGCGCCAGGCUGAAGCAGAUCGGCGUGGCUUAUUAGC__CU___CC .......................(((((......(((((...((((.....))))...)))))..)))))..((((((...................))))))................. (-20.73 = -19.38 + -1.36)
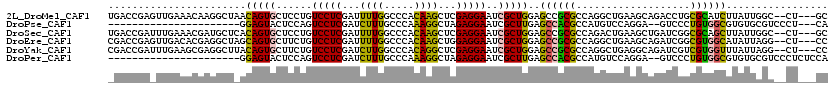
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:44 2006