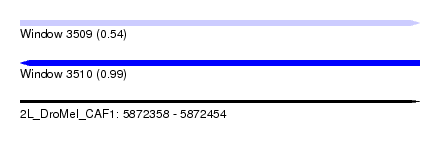
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,872,358 – 5,872,454 |
| Length | 96 |
| Max. P | 0.993419 |

| Location | 5,872,358 – 5,872,454 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 72.93 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -14.94 |
| Energy contribution | -16.00 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5872358 96 + 22407834 CAGUUCUU-----CUACUUGUG-CUGAACAGAAUCCGAGAUGUGGCCAAAUUAGGUCCAACU-UAGCCAGUUAAUGGAUUCUGUUCG-AGUUGUGCGACUACUC .(((..((-----.(((....(-((((((((((((((..(..((((.....((((.....))-))))))..)..)))))))))))).-))).))).))..))). ( -27.80) >DroSec_CAF1 62314 101 + 1 CAGGUCUUCUCAUCUACUUGUG-CUGAACAGAAUCCGAGAUUGGGCCAAAUUAGGUCCAACU-UAGCCAGUUAAUGGAUUCUGUCCG-AAUUGUGCGACUUCGC ..((((..(.((......)).)-..))((((((((((...(((((((......))))))).(-(((....)))))))))))))))).-......(((....))) ( -27.00) >DroSim_CAF1 63168 101 + 1 CAGGUCUUCUCAUCUACUUGUG-CUGAACAGAAUCCGAGAUUGGGCCAAAUUAGGUCCAACU-UAGCCAGUUAAUGGAUUCUGUCCG-AAUUGAGCGACUACGC ..((((..((((((..(....)-..))((((((((((...(((((((......))))))).(-(((....))))))))))))))...-...)))).)))).... ( -29.10) >DroEre_CAF1 86378 81 + 1 CAGGUCUU-----CAGCUUGUG-CUGAACAGAAUCCGAGAUUGGGCCAAAUUAGGUUCAACU-UAGCCAGUUAAUGGAUUCUGUCCG-A--------------- ..((...(-----((((....)-))))((((((((((...(((((((......))))))).(-(((....)))))))))))))))).-.--------------- ( -28.10) >DroYak_CAF1 87697 81 + 1 CAGGUCUU-----CUACUGCUG-CUGAACAGAAUCCGAGAUUGGGCCAAAUUAGGUUCAGCU-CAGCCAGUUAAUGGUUUCUGUACG-A--------------- (((.....-----...)))...-.((.((((((...(((.(((((((......)))))))))-).((((.....)))))))))).))-.--------------- ( -22.60) >DroAna_CAF1 60404 91 + 1 --GGUCUU-----------GGGCCAGGCCAGAAUCCGAGUUUCAGGCAAAUGGAGUUCAAAUGUGGCCUUCUAAUGGCUUCGCUCCGCAAAUGUACUACAAUAC --((..(.-----------.((((((((((.(....((..((((......))))..))...).)))))......)))))..)..)).................. ( -21.40) >consensus CAGGUCUU_____CUACUUGUG_CUGAACAGAAUCCGAGAUUGGGCCAAAUUAGGUCCAACU_UAGCCAGUUAAUGGAUUCUGUCCG_AAUUGUGCGACUAC_C .........................(.((((((((((.....(((((......)))))((((......))))..)))))))))).).................. (-14.94 = -16.00 + 1.06)

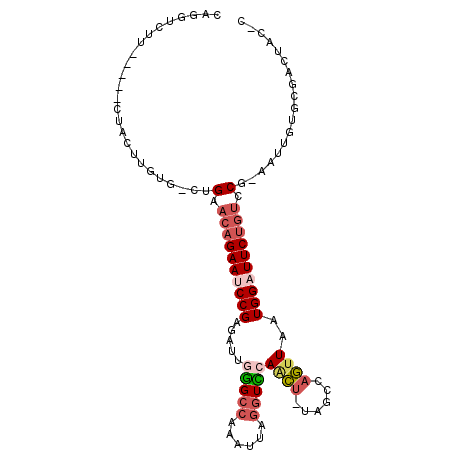
| Location | 5,872,358 – 5,872,454 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 72.93 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -19.15 |
| Energy contribution | -20.04 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5872358 96 - 22407834 GAGUAGUCGCACAACU-CGAACAGAAUCCAUUAACUGGCUA-AGUUGGACCUAAUUUGGCCACAUCUCGGAUUCUGUUCAG-CACAAGUAG-----AAGAACUG ((((.(.....).)))-)(((((((((((......((((((-(((((....)))))))))))......)))))))))))..-.........-----........ ( -34.20) >DroSec_CAF1 62314 101 - 1 GCGAAGUCGCACAAUU-CGGACAGAAUCCAUUAACUGGCUA-AGUUGGACCUAAUUUGGCCCAAUCUCGGAUUCUGUUCAG-CACAAGUAGAUGAGAAGACCUG (((....)))....((-((((((((((((.......(((((-(((((....)))))))))).......))))))))))).(-(....))......)))...... ( -32.64) >DroSim_CAF1 63168 101 - 1 GCGUAGUCGCUCAAUU-CGGACAGAAUCCAUUAACUGGCUA-AGUUGGACCUAAUUUGGCCCAAUCUCGGAUUCUGUUCAG-CACAAGUAGAUGAGAAGACCUG .....(((.((((...-.(((((((((((.......(((((-(((((....)))))))))).......))))))))))).(-(....))...))))..)))... ( -33.94) >DroEre_CAF1 86378 81 - 1 ---------------U-CGGACAGAAUCCAUUAACUGGCUA-AGUUGAACCUAAUUUGGCCCAAUCUCGGAUUCUGUUCAG-CACAAGCUG-----AAGACCUG ---------------.-.(((((((((((.......(((((-(((((....)))))))))).......)))))))))((((-(....))))-----)...)).. ( -31.34) >DroYak_CAF1 87697 81 - 1 ---------------U-CGUACAGAAACCAUUAACUGGCUG-AGCUGAACCUAAUUUGGCCCAAUCUCGGAUUCUGUUCAG-CAGCAGUAG-----AAGACCUG ---------------.-.................((.((((-.(((((((..(((((((.......)))))))..))))))-)..))))..-----.))..... ( -18.70) >DroAna_CAF1 60404 91 - 1 GUAUUGUAGUACAUUUGCGGAGCGAAGCCAUUAGAAGGCCACAUUUGAACUCCAUUUGCCUGAAACUCGGAUUCUGGCCUGGCCC-----------AAGACC-- ((.((((((.....)))))).))...((((......(((((.((((((....((......))....))))))..)))))))))..-----------......-- ( -18.30) >consensus G_GUAGUCGCACAAUU_CGGACAGAAUCCAUUAACUGGCUA_AGUUGAACCUAAUUUGGCCCAAUCUCGGAUUCUGUUCAG_CACAAGUAG_____AAGACCUG ..................(((((((((((.......(((((.(((((....)))))))))).......)))))))))))......................... (-19.15 = -20.04 + 0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:26 2006