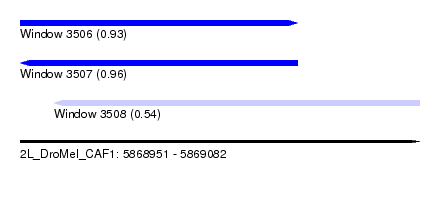
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,868,951 – 5,869,082 |
| Length | 131 |
| Max. P | 0.961230 |

| Location | 5,868,951 – 5,869,042 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -20.04 |
| Consensus MFE | -12.36 |
| Energy contribution | -13.64 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5868951 91 + 22407834 CCCAGAAAGUCCUACAUUUCUUCCGCCACUCUUUCGCCACUAAGCGAAAAAUUUAAGCAAAGUGAUACAUCUUACUUCUGGGUGAGUUGUA (((((((.................((.....((((((......)))))).......))...((((......)))))))))))......... ( -17.90) >DroSec_CAF1 58731 91 + 1 CCCACAAAGUCCCACAUUCCAUCCGCCACUUUUUCGCCACUAAGCGAAAAAUUUAAGCAAAGUGAUACAUAUUACUUCUGGGUGAGUUGUA ...((((...((((..........((...((((((((......)))))))).....)).(((((((....))))))).))))....)))). ( -20.50) >DroSim_CAF1 59575 91 + 1 CCCACAGAGUCCUACAUUCCAUCCGCCACUUUUUCGCCACUAAGCGAAAAAUUUAAGCAAAGUGAUACAUAUUACUUCUGGGUGAGUUGUA ............((((...(((((((...((((((((......)))))))).....)).(((((((....)))))))..)))))...)))) ( -20.80) >DroEre_CAF1 82862 74 + 1 CC----------CACAA------CCCCACA-CUCCGCCAGCAAGCGAAAAAUUUAAGCAAAGUGAUACAUAUUACUUUCGGGCGCGUUGUA ..----------.((((------((((...-...(((......)))..........(.((((((((....))))))))))))...))))). ( -16.40) >DroYak_CAF1 84102 82 + 1 CC---------UUACAUUUCUUCCGCCACUUUUUCGCCAGUAAGCGAAAAGUUUAGGCAAAGUGAUACAUAUUACUUUUGGGUGAAUUGUA ..---------.((((.(((.((((((..((((((((......))))))))....)))((((((((....)))))))).))).))).)))) ( -24.60) >consensus CCCA_A_AGUCCUACAUUCCAUCCGCCACUUUUUCGCCACUAAGCGAAAAAUUUAAGCAAAGUGAUACAUAUUACUUCUGGGUGAGUUGUA .......................((((..((((((((......))))))))........(((((((....)))))))...))))....... (-12.36 = -13.64 + 1.28)

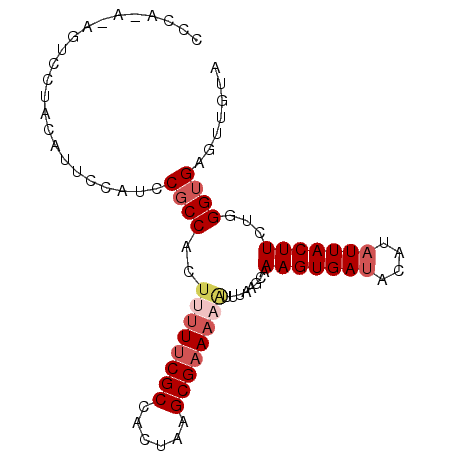
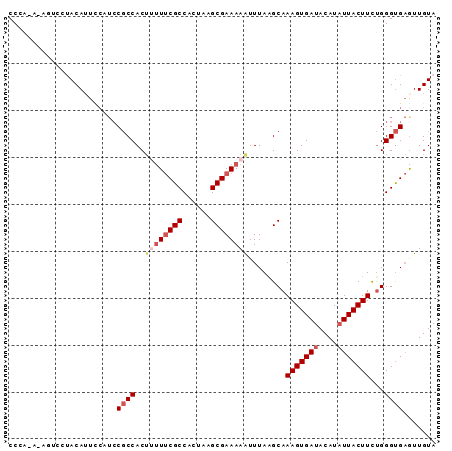
| Location | 5,868,951 – 5,869,042 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -25.86 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.46 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5868951 91 - 22407834 UACAACUCACCCAGAAGUAAGAUGUAUCACUUUGCUUAAAUUUUUCGCUUAGUGGCGAAAGAGUGGCGGAAGAAAUGUAGGACUUUCUGGG .........(((((((((...((((.((..((((((....(((((((((....)))))))))..)))))).)).))))...)).))))))) ( -29.40) >DroSec_CAF1 58731 91 - 1 UACAACUCACCCAGAAGUAAUAUGUAUCACUUUGCUUAAAUUUUUCGCUUAGUGGCGAAAAAGUGGCGGAUGGAAUGUGGGACUUUGUGGG .........(((((((((..(((((.(((.((((((....(((((((((....)))))))))..))))))))).)))))..))))).)))) ( -29.90) >DroSim_CAF1 59575 91 - 1 UACAACUCACCCAGAAGUAAUAUGUAUCACUUUGCUUAAAUUUUUCGCUUAGUGGCGAAAAAGUGGCGGAUGGAAUGUAGGACUCUGUGGG .........((((((.((..(((((.(((.((((((....(((((((((....)))))))))..))))))))).)))))..)))))).)). ( -29.40) >DroEre_CAF1 82862 74 - 1 UACAACGCGCCCGAAAGUAAUAUGUAUCACUUUGCUUAAAUUUUUCGCUUGCUGGCGGAG-UGUGGGG------UUGUG----------GG .....(((((((..((((((..((...))..)))))).....(((((((....)))))))-....)))------.))))----------.. ( -19.20) >DroYak_CAF1 84102 82 - 1 UACAAUUCACCCAAAAGUAAUAUGUAUCACUUUGCCUAAACUUUUCGCUUACUGGCGAAAAAGUGGCGGAAGAAAUGUAA---------GG ((((.(((..((.(((((.((....)).)))))(((...((((((((((....)))).)))))))))))..))).)))).---------.. ( -21.40) >consensus UACAACUCACCCAGAAGUAAUAUGUAUCACUUUGCUUAAAUUUUUCGCUUAGUGGCGAAAAAGUGGCGGAAGAAAUGUAGGACU_U_UGGG ((((..((..((.(((((..........)))))(((....(((((((((....)))))))))..)))))..))..))))............ (-15.30 = -15.46 + 0.16)

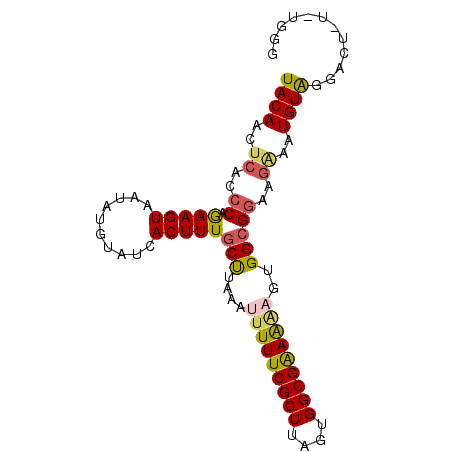
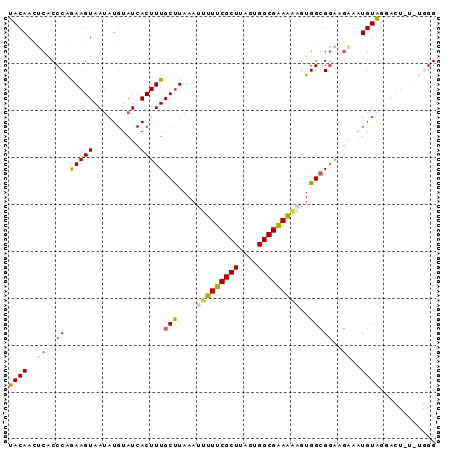
| Location | 5,868,962 – 5,869,082 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.33 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -23.74 |
| Energy contribution | -24.10 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
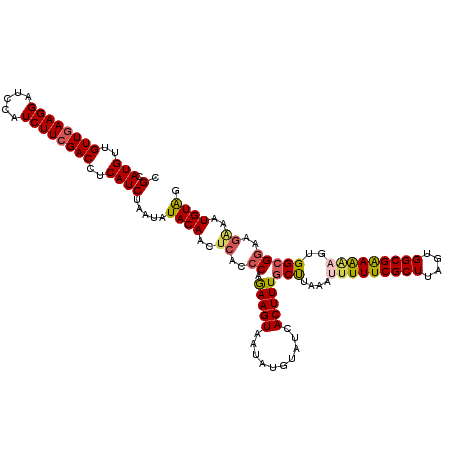
>2L_DroMel_CAF1 5868962 120 - 22407834 CGCAUGUUGUUAAAGGAUCCAUCUUCGACCUCAUCUAAUAUACAACUCACCCAGAAGUAAGAUGUAUCACUUUGCUUAAAUUUUUCGCUUAGUGGCGAAAGAGUGGCGGAAGAAAUGUAG .((((.........(....).(((((((...(((((....(((..((.....))..))))))))..))....((((....(((((((((....)))))))))..))))))))).)))).. ( -25.60) >DroSec_CAF1 58742 120 - 1 CGCAUGUUGUUGAAGGAUCCAUCUUCGACCUCAUCUAAAAUACAACUCACCCAGAAGUAAUAUGUAUCACUUUGCUUAAAUUUUUCGCUUAGUGGCGAAAAAGUGGCGGAUGGAAUGUGG (((((...((((((((.....)))))))).((((((.................(((((.((....)).)))))(((....(((((((((....)))))))))..))))))))).))))). ( -31.60) >DroSim_CAF1 59586 120 - 1 CGCAUGUUGUUGAAGGAUCCAUCUUCGACCUCAUCUAAUAUACAACUCACCCAGAAGUAAUAUGUAUCACUUUGCUUAAAUUUUUCGCUUAGUGGCGAAAAAGUGGCGGAUGGAAUGUAG .(.(((..((((((((.....))))))))..)))).....((((..(((.((.(((((.((....)).)))))(((....(((((((((....)))))))))..))))).)))..)))). ( -30.90) >DroEre_CAF1 82864 112 - 1 CGGAUGUUGUUGAAGGAUCCAUCUUCGACCUCAUCUAAUAUACAACGCGCCCGAAAGUAAUAUGUAUCACUUUGCUUAAAUUUUUCGCUUGCUGGCGGAG-UGUGGGG------UUGUG- .(((((..((((((((.....))))))))..)))))....((((((...((((.((((((..((...))..)))))).....(((((((....)))))))-..)))))------)))))- ( -32.60) >DroYak_CAF1 84104 120 - 1 CGCAUGUUGUUGAAGGAUCCAUCUUCGACCUCAUCUAAUAUACAAUUCACCCAAAAGUAAUAUGUAUCACUUUGCCUAAACUUUUCGCUUACUGGCGAAAAAGUGGCGGAAGAAAUGUAA .(.(((..((((((((.....))))))))..)))).....((((.(((..((.(((((.((....)).)))))(((...((((((((((....)))).)))))))))))..))).)))). ( -30.80) >consensus CGCAUGUUGUUGAAGGAUCCAUCUUCGACCUCAUCUAAUAUACAACUCACCCAGAAGUAAUAUGUAUCACUUUGCUUAAAUUUUUCGCUUAGUGGCGAAAAAGUGGCGGAAGAAAUGUAG .(.(((..((((((((.....))))))))..)))).....((((..((..((.(((((..........)))))(((....(((((((((....)))))))))..)))))..))..)))). (-23.74 = -24.10 + 0.36)

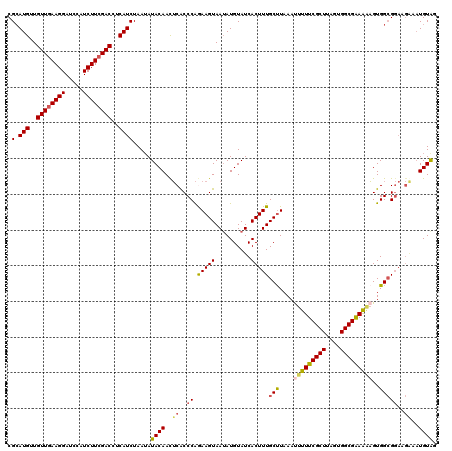
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:24 2006