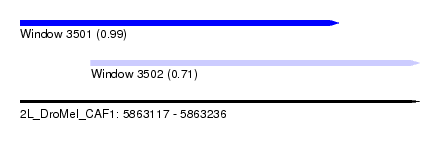
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,863,117 – 5,863,236 |
| Length | 119 |
| Max. P | 0.985244 |

| Location | 5,863,117 – 5,863,212 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.02 |
| Mean single sequence MFE | -22.12 |
| Consensus MFE | -14.49 |
| Energy contribution | -15.18 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5863117 95 + 22407834 AUAAGGUUGCAUAAAAAA-------------------GAGCGAAU-UGAGGGUCAGUGAAAAGCACUUUGCUGUCUCCAAUUGCAA-AUCAACCAAUUCCGAUCCCCAAAAAAAAA ....((((((........-------------------).((.(((-((.(((.(((..((......))..))).))))))))))..-..)))))...................... ( -20.00) >DroSec_CAF1 53093 91 + 1 AUAAGGUUGCAUAAAAAA--------------------AGCGAAUUUGAGGGUCAGUGAAAAGCACUUUGCUGUCUCCAAUUGCAAAAUCAACCAAUUCCGAGCCUCAAAA----- ......((((........--------------------.)))).(((((((.((.(((.....)))(((((...........))))).............)).))))))).----- ( -19.30) >DroSim_CAF1 53854 93 + 1 AUAAAGUUGCAUAAAAAAA------------------CAGCGAAG-UGAGGGUCAGUGAAAAGCACUUUGCUGUCUCCAAUUGCAAAAUCAACCAAUUCCGACCCUCAAAAA---- .....((((..........------------------))))....-((((((((.(((.....)))(((((...........))))).............))))))))....---- ( -21.90) >DroEre_CAF1 77318 113 + 1 AAAAAGUUGUACAAA-AAUCCUGGCAUCCAGGGCAUUGAGCGAAU-UGAGGGUCAGUGAAAAACACUUUGCUGUUUCCAAUUGCAA-AUCAACCAAUUCCAAGCACCAAAAAAAAA .....((((..(((.-..((((((...))))))..))).((.(((-((.(((.(((..((......))..))).))))))))))..-..))))....................... ( -22.90) >DroYak_CAF1 78267 114 + 1 AUAAGGUUGAACAAAAAAUCCUGGCAUUCAGAACGAUGAGCGAAU-UGAGGGUCAGUGAAAAGCACUUUGCUGUCUCCAAUUGCAA-AUCAACCAAUUUCCCCCACCAAAAAAAAA ....((((((..........(((.....)))........((.(((-((.(((.(((..((......))..))).))))))))))..-.))))))...................... ( -26.50) >consensus AUAAGGUUGCAUAAAAAA___________________GAGCGAAU_UGAGGGUCAGUGAAAAGCACUUUGCUGUCUCCAAUUGCAA_AUCAACCAAUUCCGACCCCCAAAAAAAAA ....(((((...........(((.....)))........((((...((.(((.(((..((......))..))).))))).)))).....)))))...................... (-14.49 = -15.18 + 0.69)

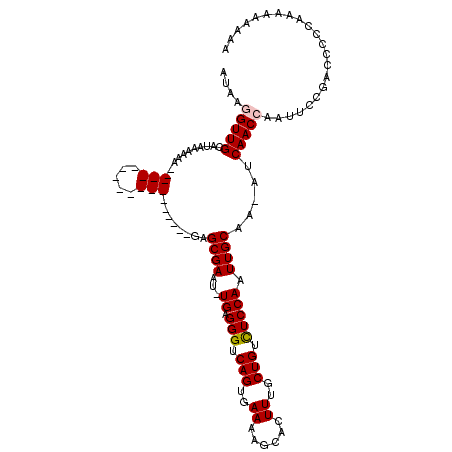
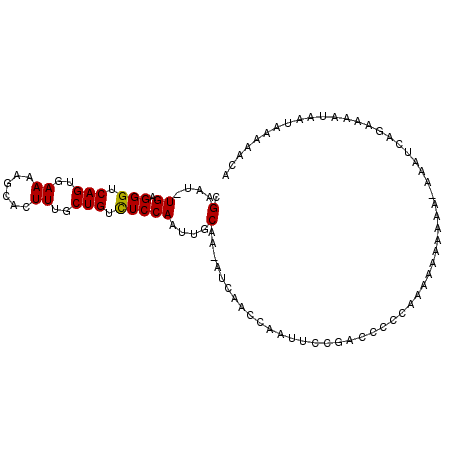
| Location | 5,863,138 – 5,863,236 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 86.78 |
| Mean single sequence MFE | -15.45 |
| Consensus MFE | -10.14 |
| Energy contribution | -9.98 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5863138 98 + 22407834 CGAAU-UGAGGGUCAGUGAAAAGCACUUUGCUGUCUCCAAUUGCAA-AUCAACCAAUUCCGAUCCCCAAAAAAAAAAAAAAUCCGAAAAUAAUAAAAACA ..(((-((.(((.(((..((......))..))).))))))))....-.........(((.(((.................))).)))............. ( -13.63) >DroSec_CAF1 53113 93 + 1 CGAAUUUGAGGGUCAGUGAAAAGCACUUUGCUGUCUCCAAUUGCAAAAUCAACCAAUUCCGAGCCUCAAAA-------AAAUCAGAAAAUAAUAAAAACA ....(((((((.((.(((.....)))(((((...........))))).............)).))))))).-------...................... ( -17.60) >DroSim_CAF1 53876 95 + 1 CGAAG-UGAGGGUCAGUGAAAAGCACUUUGCUGUCUCCAAUUGCAAAAUCAACCAAUUCCGACCCUCAAAAA----AAAAAUCAGAAAAUAAUAAAAACA .....-((((((((.(((.....)))(((((...........))))).............))))))))....----........................ ( -20.20) >DroEre_CAF1 77357 92 + 1 CGAAU-UGAGGGUCAGUGAAAAACACUUUGCUGUUUCCAAUUGCAA-AUCAACCAAUUCCAAGCACCAAAAAAAAAA------AGAAAAUAAUAAAAACA .((((-((.(((.(((..((......))..)))..)))..(((...-..))).))))))..................------................. ( -12.30) >DroYak_CAF1 78307 96 + 1 CGAAU-UGAGGGUCAGUGAAAAGCACUUUGCUGUCUCCAAUUGCAA-AUCAACCAAUUUCCCCCACCAAAAAAAAAU--AGCCAGAAAAUAAUAAAAACA ..(((-((.(((.(((..((......))..))).))))))))....-..............................--..................... ( -13.50) >consensus CGAAU_UGAGGGUCAGUGAAAAGCACUUUGCUGUCUCCAAUUGCAA_AUCAACCAAUUCCGACCCCCAAAAAAAAAA_AAAUCAGAAAAUAAUAAAAACA .(....((.(((.(((..((......))..))).)))))....)........................................................ (-10.14 = -9.98 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:19 2006