
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,824,058 – 5,824,183 |
| Length | 125 |
| Max. P | 0.717778 |

| Location | 5,824,058 – 5,824,149 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -15.00 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5824058 91 - 22407834 ACCACGAGCGGUUAUUCCGCUUA-UUCAGUACGAAAACGAGUGAAAAUAUUAUUCAAAUAUCACCCGGAGUUCUGGGUUCGAAUUCCCGCUC .....((((((..(((((((((.-(((.....)))...)))))...................((((((....))))))..))))..)))))) ( -27.30) >DroSec_CAF1 23086 91 - 1 ACCACGAGCGGUUAUUCCGCUUA-UUCAGUACGAAAACGAGUGAAAAUAUUAUUCAAAUAUCAACCGGAGUUCUGUGUUCGAAUUCCCGCUC .....((((((..(((((((..(-(((.((......))((((((.....))))))............))))...)))...))))..)))))) ( -21.50) >DroSim_CAF1 22298 90 - 1 ACCACGAGCGGUUAUUCCGCUUU-UUCAGUACGAAAACGAGUGA-AAUAUUAUUCAAAUAUCAACCGGAGUUCUAUGUCCGAAUUCCCGCUC .....((((((..((((((((((-(((.....))))..))))).-.....................(((........)))))))..)))))) ( -22.10) >DroEre_CAF1 47315 91 - 1 AUCACGAGCGGAUAGUCCGCUACUUUCAGUACGAAAACGAGUGA-AAUAUUAUUCAAAUAACACUCGGAGUUCUGGGCCCGAAUUCCCGCUC .....((((((......((.(((.....)))))....((((((.-..((((.....)))).))))))((((((.(....))))))))))))) ( -24.30) >DroYak_CAF1 47788 90 - 1 ACCACGAGCGGAUAUUCCGCUUG-UUCAGUUCGAAAACGAGUGA-AAUAUUAUUCAAAUAUCACUCGGCGUUCUGGGUCCGAGUUCCCGCUC (((((((((((.....)))))))-).(((..((....(((((((-..((((.....))))))))))).))..))))))..((((....)))) ( -31.90) >DroAna_CAF1 21115 85 - 1 AGUGCA--CGCUUGUCCGGGUAU-UUCGGUAGAAAAUCGGAUGA-AAUAUUAUCCAAA-AUCACUCGGAGCCUUGG--CCAAUUUCCCGUCC .....(--((....(((((((.(-(((....))))...((((((-....))))))...-...)))))))((....)--)........))).. ( -19.80) >consensus ACCACGAGCGGUUAUUCCGCUUA_UUCAGUACGAAAACGAGUGA_AAUAUUAUUCAAAUAUCACCCGGAGUUCUGGGUCCGAAUUCCCGCUC .....(((((............................((((((.....))))))...........(((((((.......)))))))))))) (-15.00 = -15.20 + 0.20)

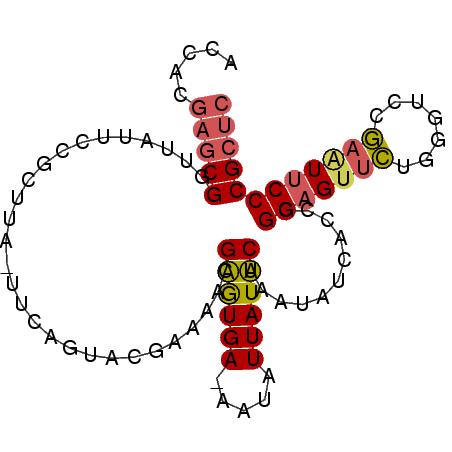
| Location | 5,824,086 – 5,824,183 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.43 |
| Mean single sequence MFE | -21.63 |
| Consensus MFE | -14.28 |
| Energy contribution | -14.78 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5824086 97 - 22407834 UCUGUCGAAUCCGAACAUUCGGC----GAACUUACACAACCACGAGCGGUUAUUCCGCUUA-UUCAGUACGAAAACGAGUGAAAAUAUUAUUCAAAUAUCAC ...(((((((......)))))))----(((.............((((((.....)))))).-((((.(.((....))).)))).......)))......... ( -20.00) >DroSec_CAF1 23114 97 - 1 CCUGUCGAAUCCGAACAUUCGGC----GAACUUACACAACCACGAGCGGUUAUUCCGCUUA-UUCAGUACGAAAACGAGUGAAAAUAUUAUUCAAAUAUCAA ...(((((((......)))))))----(((.............((((((.....)))))).-((((.(.((....))).)))).......)))......... ( -20.00) >DroSim_CAF1 22326 96 - 1 CCUGUAGAAUCCAAACAUUCGGC----GAACUUACACAACCACGAGCGGUUAUUCCGCUUU-UUCAGUACGAAAACGAGUGA-AAUAUUAUUCAAAUAUCAA ......((((.....((((((..----................((((((.....))))))(-(((.....)))).)))))).-......))))......... ( -16.90) >DroEre_CAF1 47343 97 - 1 CCUGUCGAAUCCGCGCAUUCGGC----GAACUUACACAAUCACGAGCGGAUAGUCCGCUACUUUCAGUACGAAAACGAGUGA-AAUAUUAUUCAAAUAACAC ..((((((((......)))))))----)...........((((.(((((.....)))))..((((.....))))....))))-................... ( -20.70) >DroYak_CAF1 47816 96 - 1 CCUGUCGAAUCCGCGCAUUCGGC----GAACUUACACAACCACGAGCGGAUAUUCCGCUUG-UUCAGUUCGAAAACGAGUGA-AAUAUUAUUCAAAUAUCAC ......((((.....((((((.(----(((((.........((((((((.....)))))))-)..))))))....)))))).-......))))......... ( -26.50) >DroAna_CAF1 21141 97 - 1 CAUGUCGAUGCGAAGCAAUCGACAGCCAAACUCACACAAGUGCA--CGCUUGUCCGGGUAU-UUCGGUAGAAAAUCGGAUGA-AAUAUUAUCCAAA-AUCAC ..(((((((((...)).)))))))(((..((((..((((((...--.))))))..))))..-...)))........((((((-....))))))...-..... ( -25.70) >consensus CCUGUCGAAUCCGAACAUUCGGC____GAACUUACACAACCACGAGCGGUUAUUCCGCUUA_UUCAGUACGAAAACGAGUGA_AAUAUUAUUCAAAUAUCAC ...(((((((......)))))))....................((((((.....))))))................((((((.....))))))......... (-14.28 = -14.78 + 0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:10 2006