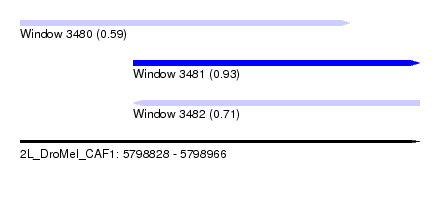
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,798,828 – 5,798,966 |
| Length | 138 |
| Max. P | 0.934070 |

| Location | 5,798,828 – 5,798,942 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 94.19 |
| Mean single sequence MFE | -23.33 |
| Consensus MFE | -19.89 |
| Energy contribution | -19.67 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5798828 114 + 22407834 CCAAGAAUGUUAUCAAUCUUAUCAUUCAAGUGGUAUAG-AACCGCAUGAUUAGCGCUCAAGGAAAACCCCAUAAAUGACGACAAUCAUGGGAAAUUUUAAACAAAUUAUGGUAAC ........(((((((.(((((((((....)))))).))-).((.(((((((..((.(((.((......)).....)))))..))))))))).................))))))) ( -23.70) >DroSec_CAF1 36050 115 + 1 CCAAGAAUGUUAUCAAUCUUAUUAUUCUAGUGGUCUAGGAACCGCAUGAUUAGCGUCCAAGGAAAAACCCAUAGAUGACGACAAUCAUGGGAAAUUUUAAACAAAUUAUAGAAAC ..((((.((....)).))))....((((((....)))))).((.(((((((..((((...((......))......))))..)))))))))........................ ( -22.90) >DroSim_CAF1 38475 114 + 1 CCAAGAAUGUUAUCAAUCUUAUCAUUCUAGUGGUCUAG-AACCGCAUGAUUAGCGUCCAAGGAAAAACCCAUAAAUGACGACAAUCAUGGGAAAUUUUAAACAAAUUAUAGAAAC ..((((.((....)).)))).((.((((((....))))-))((.(((((((..((((...((......))......))))..)))))))))...................))... ( -23.40) >consensus CCAAGAAUGUUAUCAAUCUUAUCAUUCUAGUGGUCUAG_AACCGCAUGAUUAGCGUCCAAGGAAAAACCCAUAAAUGACGACAAUCAUGGGAAAUUUUAAACAAAUUAUAGAAAC (((.((.(((..(((..((.(((((....(((((......)))))))))).)).......((......)).....)))..))).)).)))......................... (-19.89 = -19.67 + -0.22)

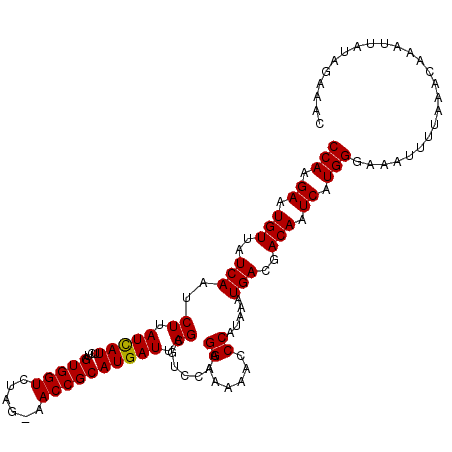
| Location | 5,798,867 – 5,798,966 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.61 |
| Mean single sequence MFE | -16.11 |
| Consensus MFE | -14.27 |
| Energy contribution | -14.17 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5798867 99 + 22407834 ACCGCAUGAUUAGCGCUCAAGGAAAACCCCAUAAAUGACGACAAUCAUGGGAAAUUUUAAACAAAUUAUGGUAACAUUAUAAACCUUAAAAAU----UGUACU .((.(((((((..((.(((.((......)).....)))))..))))))))).........((((.(((.(((..........))).)))...)----)))... ( -15.60) >DroSec_CAF1 36090 86 + 1 ACCGCAUGAUUAGCGUCCAAGGAAAAACCCAUAGAUGACGACAAUCAUGGGAAAUUUUAAACAAAUUAUAGAAACAUUAUAAAUC-----------------U .((.(((((((..((((...((......))......))))..)))))))))..................................-----------------. ( -15.90) >DroSim_CAF1 38514 103 + 1 ACCGCAUGAUUAGCGUCCAAGGAAAAACCCAUAAAUGACGACAAUCAUGGGAAAUUUUAAACAAAUUAUAGAAACAUUAUAAAUCUUAAAAAUUUAUUGAUCU .((.(((((((..((((...((......))......))))..)))))))))(((((((.......((((((.....))))))......)))))))........ ( -16.82) >consensus ACCGCAUGAUUAGCGUCCAAGGAAAAACCCAUAAAUGACGACAAUCAUGGGAAAUUUUAAACAAAUUAUAGAAACAUUAUAAAUCUUAAAAAU____UG__CU .((.(((((((..((((...((......))......))))..)))))))))..............((((((.....))))))..................... (-14.27 = -14.17 + -0.11)

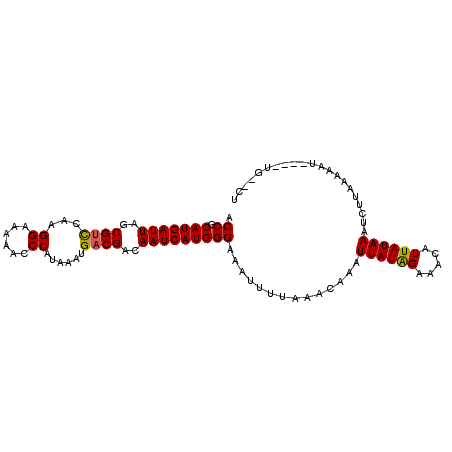
| Location | 5,798,867 – 5,798,966 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.61 |
| Mean single sequence MFE | -19.40 |
| Consensus MFE | -16.62 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5798867 99 - 22407834 AGUACA----AUUUUUAAGGUUUAUAAUGUUACCAUAAUUUGUUUAAAAUUUCCCAUGAUUGUCGUCAUUUAUGGGGUUUUCCUUGAGCGCUAAUCAUGCGGU ......----...(((((((................(((((.....)))))((((((((.((....)).))))))))....)))))))(((.......))).. ( -19.00) >DroSec_CAF1 36090 86 - 1 A-----------------GAUUUAUAAUGUUUCUAUAAUUUGUUUAAAAUUUCCCAUGAUUGUCGUCAUCUAUGGGUUUUUCCUUGGACGCUAAUCAUGCGGU .-----------------..................................((((((((((.((((......(((....)))...)))).)))))))).)). ( -18.30) >DroSim_CAF1 38514 103 - 1 AGAUCAAUAAAUUUUUAAGAUUUAUAAUGUUUCUAUAAUUUGUUUAAAAUUUCCCAUGAUUGUCGUCAUUUAUGGGUUUUUCCUUGGACGCUAAUCAUGCGGU (((.(((((((((.....)))))))..))..)))..................((((((((((.((((......(((....)))...)))).)))))))).)). ( -20.90) >consensus AG__CA____AUUUUUAAGAUUUAUAAUGUUUCUAUAAUUUGUUUAAAAUUUCCCAUGAUUGUCGUCAUUUAUGGGUUUUUCCUUGGACGCUAAUCAUGCGGU ....................................................((((((((((.((((......(((.....)))..)))).)))))))).)). (-16.62 = -16.73 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:00 2006