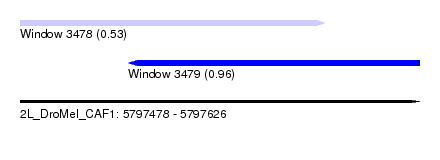
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,797,478 – 5,797,626 |
| Length | 148 |
| Max. P | 0.964253 |

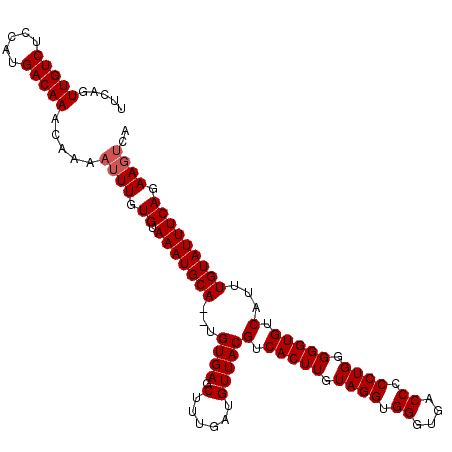
| Location | 5,797,478 – 5,797,591 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 95.85 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -28.63 |
| Energy contribution | -29.30 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5797478 113 + 22407834 UUCAGUUGUCUCCAUGACAAACAAAAUUUGUGUAAAUGCAUAUGUGAGCUUUGAAGUUACGUCACUUGUAGGUGGGUGACCCCCUGGGGGUGUCAUUUGUAUUUCAGAAGUCA .....(((((.....))))).....((((.((.(((((((...((((.((....))))))(.(((((.((((.((....)).)))).))))).)...))))))))).)))).. ( -33.00) >DroSec_CAF1 34699 111 + 1 UUCAGUUGUCUCCAUGACAAACAAAAUUUGUGUAAAUGCA--UGUGAGCUUUGAUGUUACGUCACUUGUAGGUGGGUGACCCCCUGGGGGUGUCAUUUGUAUUUCAGAAGUCA .....(((((.....))))).....((((.((.(((((((--.((((((......)).....(((((.((((.((....)).)))).))))))))).))))))))).)))).. ( -33.00) >DroSim_CAF1 37139 107 + 1 UUCAGUUGUCUCCAUGACAAACAAAAUUUGUGUAAAUGCA--UGUGAGCUUUGAUGUUACGUCACUUGUAGGUGGGUGACCCCCUGGGGGUGUCAUUUGUAUUUCAGAA---- .....(((((.....))))).......((.((.(((((((--.((((((......)).....(((((.((((.((....)).)))).))))))))).))))))))).))---- ( -30.80) >consensus UUCAGUUGUCUCCAUGACAAACAAAAUUUGUGUAAAUGCA__UGUGAGCUUUGAUGUUACGUCACUUGUAGGUGGGUGACCCCCUGGGGGUGUCAUUUGUAUUUCAGAAGUCA .....(((((.....))))).....((((.((.(((((((...((((.(......)))))(.(((((.((((.(((...))))))).))))).)...))))))))).)))).. (-28.63 = -29.30 + 0.67)


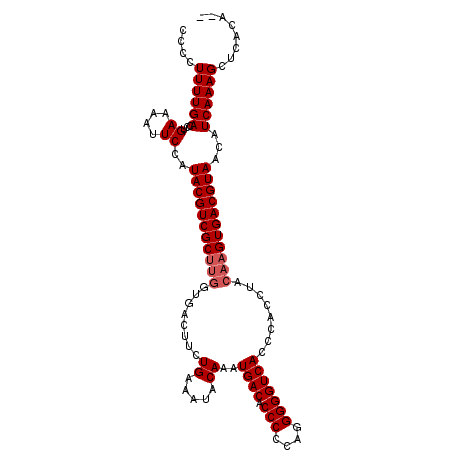
| Location | 5,797,518 – 5,797,626 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 92.55 |
| Mean single sequence MFE | -25.93 |
| Consensus MFE | -22.80 |
| Energy contribution | -23.80 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5797518 108 - 22407834 CCCCUUUUGACCUGAAAAAUCCAUACGUCGCUUGGUGACUUCUGAAAUACAAAUGACACCCCCAGGGGGUCACCCACCUACAAGUGACGUAACUUCAAAGCUCACAUA ....((((((...((....))..((((((((((((((.(....).........((((.(((....)))))))..))))...))))))))))...))))))........ ( -27.60) >DroSec_CAF1 34739 106 - 1 CCCCUUUUGACCUGAAAAUUCCAUACGUCGCUUGGUGACUUCUGAAAUACAAAUGACACCCCCAGGGGGUCACCCACCUACAAGUGACGUAACAUCAAAGCUCACA-- ....((((((...((....))..((((((((((((((.(....).........((((.(((....)))))))..))))...))))))))))...))))))......-- ( -27.70) >DroSim_CAF1 37179 98 - 1 CCCCUUUUGACCUGAAAAUUCCAUACGUCGC--------UUCUGAAAUACAAAUGACACCCCCAGGGGGUCACCCACCUACAAGUGACGUAACAUCAAAGCUCACA-- ....((((((...((....))..((((((((--------((............((((.(((....))))))).........))))))))))...))))))......-- ( -22.50) >consensus CCCCUUUUGACCUGAAAAUUCCAUACGUCGCUUGGUGACUUCUGAAAUACAAAUGACACCCCCAGGGGGUCACCCACCUACAAGUGACGUAACAUCAAAGCUCACA__ ....((((((...((....))..(((((((((((........((.....))..((((.(((....)))))))........)))))))))))...))))))........ (-22.80 = -23.80 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:57 2006