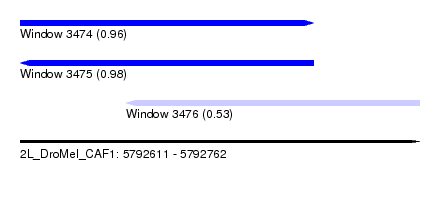
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,792,611 – 5,792,762 |
| Length | 151 |
| Max. P | 0.980156 |

| Location | 5,792,611 – 5,792,722 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -22.76 |
| Energy contribution | -23.43 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5792611 111 + 22407834 GAACACAUUUUUUGAGGGAAAACUUUUCGUUUUCCUAAUUCACGGCUGUCACUUUUCGCGUCGAGCGGGACGAAGAAAAAAAAACGUGACAAGCUGACGUUAUCAAUAAAA .......(((.((((((((((((.....))))))))......(((((((((((((((.((((......))))..)))))......))))).)))))......)))).))). ( -30.80) >DroSec_CAF1 29697 96 + 1 GGAUACAUUUUUUGAGGGAAAACUUUUCGUUUUCCUAAUUCACGCCUGUCACUUUUCGCGUCAAGCGGGACGAAGUAAAAAA-AAGUGACAAGCUGA-------------- ...............((((((((.....))))))))...(((.((.((((((((((..((((......))))........))-)))))))).)))))-------------- ( -27.50) >DroSim_CAF1 32224 107 + 1 GGAUACAUUAUUUGAGGGAAAACUUUUCGUUUUCCUAAUUCACGCCUGUCACUUUUCCCGUCAAGCGGGACGAAG---AAAA-AAGUGACAAGCUGACGUUAUCAAUAAAA .(((((.........((((((((.....))))))))...(((.((.((((((((((((((.....))))).....---...)-)))))))).))))).).))))....... ( -33.80) >consensus GGAUACAUUUUUUGAGGGAAAACUUUUCGUUUUCCUAAUUCACGCCUGUCACUUUUCGCGUCAAGCGGGACGAAG_AAAAAA_AAGUGACAAGCUGACGUUAUCAAUAAAA ...............((((((((.....))))))))...(((.((.((((((((....((((......))))...........)))))))).))))).............. (-22.76 = -23.43 + 0.67)

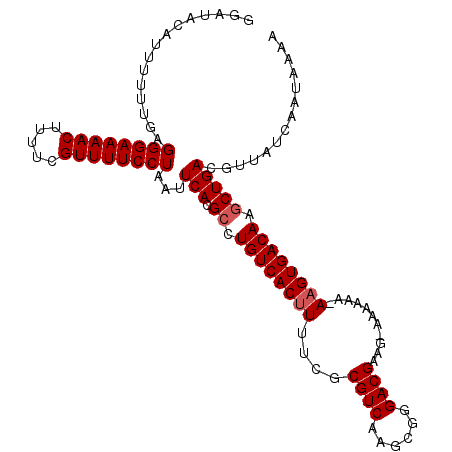
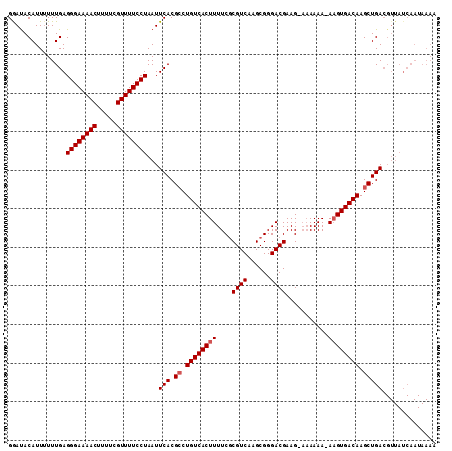
| Location | 5,792,611 – 5,792,722 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -24.29 |
| Energy contribution | -24.96 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5792611 111 - 22407834 UUUUAUUGAUAACGUCAGCUUGUCACGUUUUUUUUUCUUCGUCCCGCUCGACGCGAAAAGUGACAGCCGUGAAUUAGGAAAACGAAAAGUUUUCCCUCAAAAAAUGUGUUC .....((((..(((...(((.(((((......(((((..((((......)))).))))))))))))))))......(((((((.....))))))).))))........... ( -24.90) >DroSec_CAF1 29697 96 - 1 --------------UCAGCUUGUCACUU-UUUUUUACUUCGUCCCGCUUGACGCGAAAAGUGACAGGCGUGAAUUAGGAAAACGAAAAGUUUUCCCUCAAAAAAUGUAUCC --------------((((((((((((((-(((.......((((......)))).)))))))))))))).)))....(((((((.....)))))))................ ( -30.90) >DroSim_CAF1 32224 107 - 1 UUUUAUUGAUAACGUCAGCUUGUCACUU-UUUU---CUUCGUCCCGCUUGACGGGAAAAGUGACAGGCGUGAAUUAGGAAAACGAAAAGUUUUCCCUCAAAUAAUGUAUCC .....((((.....((((((((((((((-((..---(.(((.......))).)..))))))))))))).)))....(((((((.....))))))).))))........... ( -36.40) >consensus UUUUAUUGAUAACGUCAGCUUGUCACUU_UUUUUU_CUUCGUCCCGCUUGACGCGAAAAGUGACAGGCGUGAAUUAGGAAAACGAAAAGUUUUCCCUCAAAAAAUGUAUCC ..............((((((((((((((...........((((......))))....))))))))))).)))....(((((((.....)))))))................ (-24.29 = -24.96 + 0.67)

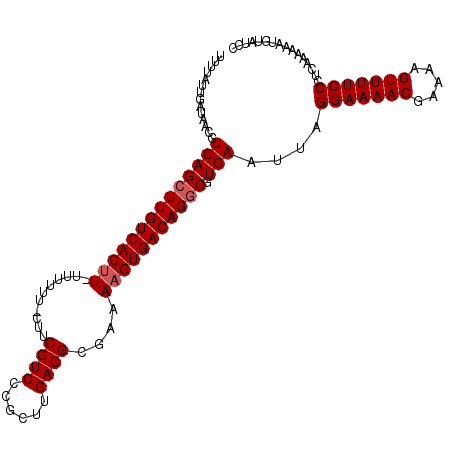
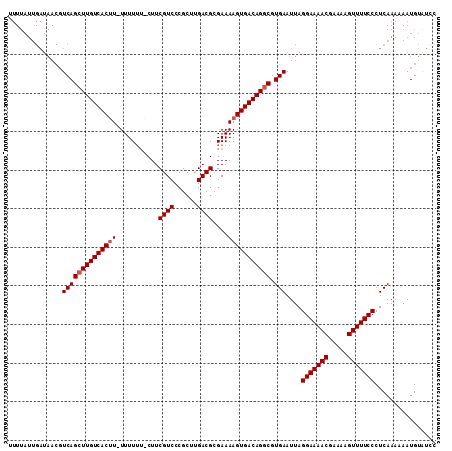
| Location | 5,792,651 – 5,792,762 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 84.04 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -18.79 |
| Energy contribution | -19.79 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5792651 111 - 22407834 UAUUCGAUUUUAUAUUGAACUCAAGUGCGCUUUCAGUGUGUUUUAUUGAUAACGUCAGCUUGUCACGUUUUUUUUUCUUCGUCCCGCUCGACGCGAAAAGUGACAGCCGUG ...(((((..(((((((((..(......)..)))))))))....)))))..(((...(((.(((((......(((((..((((......)))).)))))))))))))))). ( -23.90) >DroSec_CAF1 29737 94 - 1 AAUUCGAUUUUAUAUUGAACUCAAGUGCGCUUUCAGUG----------------UCAGCUUGUCACUU-UUUUUUACUUCGUCCCGCUUGACGCGAAAAGUGACAGGCGUG ..((((((.....)))))).......((((.....)))----------------)..(((((((((((-(((.......((((......)))).))))))))))))))... ( -26.40) >DroSim_CAF1 32264 107 - 1 UAUUCGAUUUUAUAUUGAACUCAAGUGCGCUCUCAGUGUGUUUUAUUGAUAACGUCAGCUUGUCACUU-UUUU---CUUCGUCCCGCUUGACGGGAAAAGUGACAGGCGUG ..((((((.....)))))).((((..((((.......))))....))))........(((((((((((-((..---(.(((.......))).)..)))))))))))))... ( -33.30) >consensus UAUUCGAUUUUAUAUUGAACUCAAGUGCGCUUUCAGUGUGUUUUAUUGAUAACGUCAGCUUGUCACUU_UUUUUU_CUUCGUCCCGCUUGACGCGAAAAGUGACAGGCGUG ..((((((.....)))))).......((((.....))))..................(((((((((((...........((((......))))....)))))))))))... (-18.79 = -19.79 + 1.00)

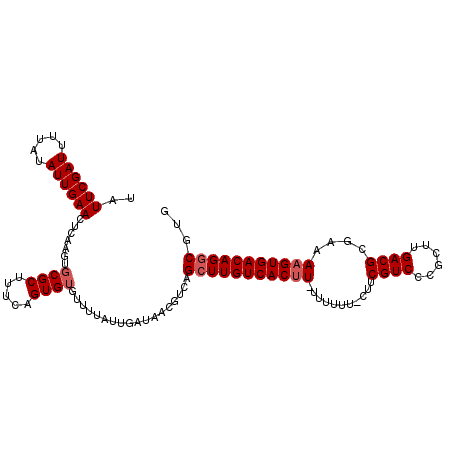
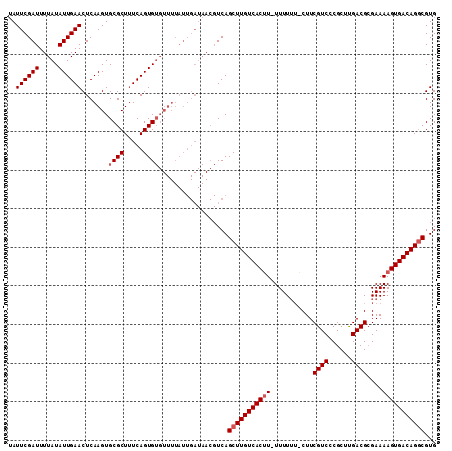
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:54 2006