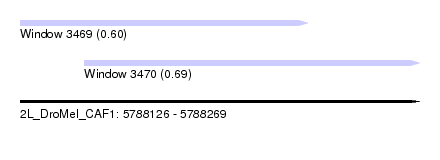
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,788,126 – 5,788,269 |
| Length | 143 |
| Max. P | 0.690865 |

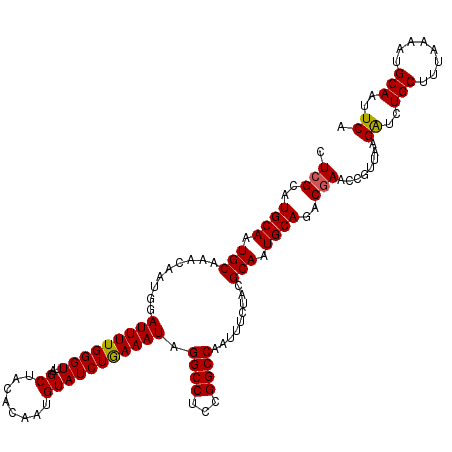
| Location | 5,788,126 – 5,788,229 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.67 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -14.75 |
| Energy contribution | -14.53 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5788126 103 + 22407834 UUCGUAUUUUAGCCUAAAGCAAG-----------------CUAGAAUGCAAUGCAAACAAUGGAUUUUGGGUUAGCUACACAAUGUAUCUAAAAUAGGCCUCCGGCCAAUUUCUACGCAU ...((((((((((.........)-----------------))))))))).((((.........(((((((((..((........))))))))))).((((...)))).........)))) ( -26.30) >DroSec_CAF1 25167 120 + 1 UCCGUAUUUUCGCCAAAAGCAAGCUAAGAUCCCCAGCACUCUCGCAUGCAAUGCAAACAAUGGAUUUUGGGUUAGCUACACAAUGUAUCUGAAAUAGGCCUCCGGCCAAUUUCUACGCAA ..((((...........(((((.(((((((((...((......)).(((...)))......))))))))).)).))).............(((((.((((...)))).)))))))))... ( -28.80) >DroSim_CAF1 27273 120 + 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNAGCACUCUCGCAUGCAAUGCAAACAAUGGAUUUUGGGUUAGCUACACAAUGUAUCUGAAAUAGGCCUCCGGCCAAUUUCUACGCAA ...................................(((........)))..(((.......((((.(((.((.....)).)))...))))(((((.((((...)))).)))))...))). ( -18.40) >consensus U_CGUAUUUU_GCC_AAAGCAAG___________AGCACUCUCGCAUGCAAUGCAAACAAUGGAUUUUGGGUUAGCUACACAAUGUAUCUGAAAUAGGCCUCCGGCCAAUUUCUACGCAA ...................................................(((.........(((((((((..((........))))))))))).((((...)))).........))). (-14.75 = -14.53 + -0.22)

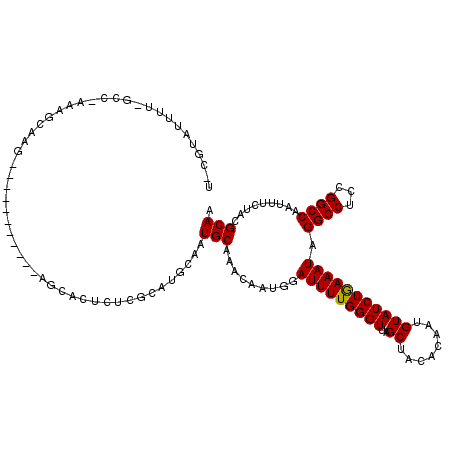
| Location | 5,788,149 – 5,788,269 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -30.93 |
| Consensus MFE | -27.31 |
| Energy contribution | -27.20 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5788149 120 + 22407834 CUAGAAUGCAAUGCAAACAAUGGAUUUUGGGUUAGCUACACAAUGUAUCUAAAAUAGGCCUCCGGCCAAUUUCUACGCAUUGCAGACGAACCGUUAAGGUCUGCUUUAAAAUGCAAUUCA ...(((((((((((.........(((((((((..((........))))))))))).((((...)))).........)))))((((((...........))))))........)).)))). ( -30.60) >DroSec_CAF1 25207 120 + 1 CUCGCAUGCAAUGCAAACAAUGGAUUUUGGGUUAGCUACACAAUGUAUCUGAAAUAGGCCUCCGGCCAAUUUCUACGCAAUGCAGACGAACCGUUAAGAUCUGCUUUAAACUGCAAUUCA .(((..((((.(((.......((((.(((.((.....)).)))...))))(((((.((((...)))).)))))...))).))))..)))........((..(((........)))..)). ( -31.10) >DroSim_CAF1 27313 120 + 1 CUCGCAUGCAAUGCAAACAAUGGAUUUUGGGUUAGCUACACAAUGUAUCUGAAAUAGGCCUCCGGCCAAUUUCUACGCAAUGCAGACGAACCGUUAAGAUCUGCUUUAAAAUGCAAUUCA .(((..((((.(((.......((((.(((.((.....)).)))...))))(((((.((((...)))).)))))...))).))))..)))........((..(((........)))..)). ( -31.10) >consensus CUCGCAUGCAAUGCAAACAAUGGAUUUUGGGUUAGCUACACAAUGUAUCUGAAAUAGGCCUCCGGCCAAUUUCUACGCAAUGCAGACGAACCGUUAAGAUCUGCUUUAAAAUGCAAUUCA .(((..((((.(((.........(((((((((..((........))))))))))).((((...)))).........))).))))..)))........((..(((........)))..)). (-27.31 = -27.20 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:48 2006