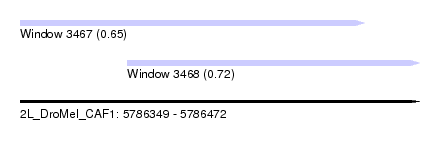
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,786,349 – 5,786,472 |
| Length | 123 |
| Max. P | 0.719763 |

| Location | 5,786,349 – 5,786,455 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -7.82 |
| Energy contribution | -9.82 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5786349 106 + 22407834 GAAUGUGUGCGAAU---GAGUAU--AAAA--CAAUACAUUCG-----AAAGCAAAC-CGAAAGUCAUUGCG-AGAGAAAUCUGUGCAAAUGAAGAAAAGGUGUCAAGAACUUUAUUGACA ...(((...(((((---(.((..--....--..)).))))))-----...))).((-(.....((((((((-(((....))).))).)))))......)))(((((........))))). ( -27.10) >DroVir_CAF1 35269 111 + 1 GUGUGUGCUAAGAG---AAAUAA--AAAG--GAAAACCCUCGAACAGAAAGCAAAG-UUAAAGUCAUGUCG-AGAGAAAUCUGAGCAAAUGAAGUAAAGGUGUCAAGAACUUUAUUGACA .....((((.(((.---......--...(--(....))(((((.((((.(((...)-))....)).)))))-)).....))).)))).............((((((........)))))) ( -17.30) >DroSec_CAF1 23427 106 + 1 GAAUGUUUUCGAAU---GAGUAU--AAAA--CAAUACAGUCG-----AAAGCAAAC-CGAAAGUCAUUGCG-AGAGAAAUCUGUGCAAAUGAAGAAAAGGUGUCAAGAACUUUAUUGACA ...(((((((((..---..((((--....--..))))..)))-----)))))).((-(.....((((((((-(((....))).))).)))))......)))(((((........))))). ( -30.20) >DroSim_CAF1 24201 106 + 1 GAAUGUGUUCGAAU---GAGUAU--AAAA--CAAUACAGUCG-----AAAGCAAAC-CGAAAGUCAUUGCG-AGAGAAAUCUGUGCAAAUGAAGAAAAGGUGUCAAGAACUUUAUUGACA ...(((.(((((..---..((((--....--..))))..)))-----)).))).((-(.....((((((((-(((....))).))).)))))......)))(((((........))))). ( -27.00) >DroWil_CAF1 44524 108 + 1 GCAUGUGCUAAAAGCCAAAGUAU--AAAAAACGAAGCAU----------GGCAAAUCUUAAAGUCAUGCCAAAGAGAAAUCUGAGCAAAUGAAGUAAAGGUGUCAAGAACUUUAUUGACA .(((.((((..............--..........((((----------(((..........)))))))...(((....))).)))).))).........((((((........)))))) ( -22.50) >DroMoj_CAF1 42969 113 + 1 GUGUUUGUUAAGUA---AAAUAAAAAAAG--GAAAACCCUCGAACAGAAAGCAAUG-CUAAAGUCAUGUCG-AGAGAAAUCUGAGCAAAUGAAGUAAAGGUGUCAAGAACUUUAUUGACA ..(((((((.((..---...........(--(....))(((((.((((.(((...)-))....)).)))))-))......)).)))))))..........((((((........)))))) ( -18.10) >consensus GAAUGUGUUAAAAU___AAGUAU__AAAA__CAAAACACUCG_____AAAGCAAAC_CGAAAGUCAUGGCG_AGAGAAAUCUGAGCAAAUGAAGAAAAGGUGUCAAGAACUUUAUUGACA ...............................................................((((.....(((....)))......))))........((((((........)))))) ( -7.82 = -9.82 + 2.00)


| Location | 5,786,382 – 5,786,472 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -13.36 |
| Energy contribution | -16.80 |
| Covariance contribution | 3.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5786382 90 + 22407834 CG-----AAAGCAAAC-CGAAAGUCAUUGCG-AGAGAAAUCUGUGCAAAUGAAGAAAAGGUGUCAAGAACUUUAUUGACAAGCAGUCCUCAAGCUGC--- ..-----.......((-(.....((((((((-(((....))).))).)))))......)))(((((........)))))..(((((......)))))--- ( -23.40) >DroVir_CAF1 35302 90 + 1 CGAACAGAAAGCAAAG-UUAAAGUCAUGUCG-AGAGAAAUCUGAGCAAAUGAAGUAAAGGUGUCAAGAACUUUAUUGACAGG----CUUCAAGCUG---- .........(((....-.........(((..-(((....)))..)))..((((((.....((((((........)))))).)----))))).))).---- ( -20.30) >DroSec_CAF1 23460 90 + 1 CG-----AAAGCAAAC-CGAAAGUCAUUGCG-AGAGAAAUCUGUGCAAAUGAAGAAAAGGUGUCAAGAACUUUAUUGACAAGCAGUCCUCAAGCUGC--- ..-----.......((-(.....((((((((-(((....))).))).)))))......)))(((((........)))))..(((((......)))))--- ( -23.40) >DroSim_CAF1 24234 90 + 1 CG-----AAAGCAAAC-CGAAAGUCAUUGCG-AGAGAAAUCUGUGCAAAUGAAGAAAAGGUGUCAAGAACUUUAUUGACAAGCAGUCCUCAAGCUGC--- ..-----.......((-(.....((((((((-(((....))).))).)))))......)))(((((........)))))..(((((......)))))--- ( -23.40) >DroWil_CAF1 44561 87 + 1 ---------GGCAAAUCUUAAAGUCAUGCCAAAGAGAAAUCUGAGCAAAUGAAGUAAAGGUGUCAAGAACUUUAUUGACAGG----CCUCAAGCUGCUGG ---------(((..(((((.(..((((((...(((....)))..)))..)))..).)))))(((((........)))))..)----))............ ( -20.50) >consensus CG_____AAAGCAAAC_CGAAAGUCAUUGCG_AGAGAAAUCUGUGCAAAUGAAGAAAAGGUGUCAAGAACUUUAUUGACAAGCAGUCCUCAAGCUGC___ .........(((..............(((((.(((....))).))))).(((.((.....((((((........)))))).....)).))).)))..... (-13.36 = -16.80 + 3.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:47 2006