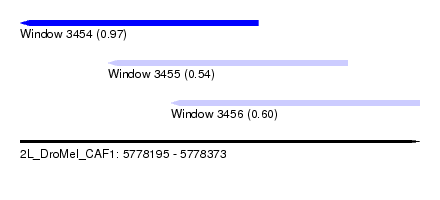
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,778,195 – 5,778,373 |
| Length | 178 |
| Max. P | 0.970880 |

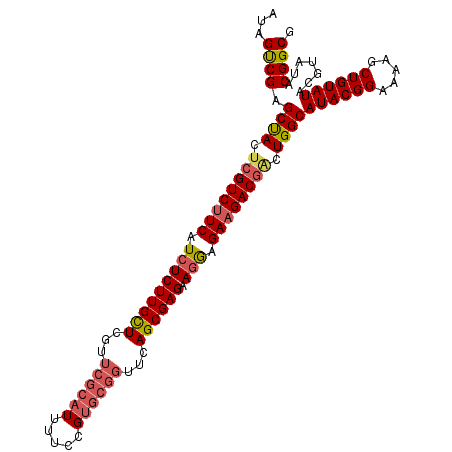
| Location | 5,778,195 – 5,778,301 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 90.65 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -35.69 |
| Energy contribution | -37.70 |
| Covariance contribution | 2.01 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5778195 106 - 22407834 AUAGUCGAGCCACUAGUC-UCAUAUCUUGCUCGUUCGCAUUUUUCGUGCGGUUCAGCGAGAAGAAGAAGACGGCUGGCAUACGGAAAAGCUGUAUACGUAUACGGCG ...((((.((((...(((-((...(((((((...((((((.....))))))...)))))))....).))))...))))((((((.....)))))).......)))). ( -35.90) >DroSec_CAF1 15298 102 - 1 AUAGUCGAGCUACUCGUCUUCAUCUCUUGUUCGUUCGCAUUUUGCG-----UUCAGCGAGAAGGAGAAGACGACUGGCAUACGGAAAAGCUGUAUACGUAUACGGCG ...((((.((((.((((((((..((((((((.(..(((.....)))-----..))))))).))..)))))))).))))((((((.....)))))).......)))). ( -41.00) >DroSim_CAF1 16062 107 - 1 AUAGCCGAGCUACUCGUCUUCAUCUCUUGCUCGUUCGCAUUUUCCGUGCGGUUCAGCGAGAAGGAGAAGACGUCUGGCAUACGGAAAAGCUGUAUACGUAUACGGCG ...((((.((((..(((((((..((((((((...((((((.....))))))...)))))).))..)))))))..))))((((((.....)))))).......)))). ( -46.40) >consensus AUAGUCGAGCUACUCGUCUUCAUCUCUUGCUCGUUCGCAUUUUCCGUGCGGUUCAGCGAGAAGGAGAAGACGACUGGCAUACGGAAAAGCUGUAUACGUAUACGGCG ...((((.((((.((((((((.(((((((((...((((((.....))))))...)))))).))).)))))))).))))((((((.....)))))).......)))). (-35.69 = -37.70 + 2.01)

| Location | 5,778,234 – 5,778,341 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -19.18 |
| Energy contribution | -21.30 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5778234 107 - 22407834 UAGUGGAACUGUUGCUCAAAGUGCGAGCAAUGAGUAUCUGAUAGUCGAGCCACUAGUC-UCAUAUCUUGCUCGUUCGCAUUUUUCGUGCGGUUCAGCGAGAAGAAGAA ..((.(((((((.....(((((((((((...(((((...((((...(((.(....).)-)).)))).))))))))))))))))....))))))).))........... ( -33.70) >DroSec_CAF1 15337 91 - 1 ------------UGCUCAAAUUGCGAGUAGUGGGUAUCUGAUAGUCGAGCUACUCGUCUUCAUCUCUUGUUCGUUCGCAUUUUGCG-----UUCAGCGAGAAGGAGAA ------------..........(((((((((((.(((...))).))..))))))))).....((((((.(((((((((.....)))-----...)))))).)))))). ( -27.40) >DroSim_CAF1 16101 96 - 1 ------------UGCUAAAAUUGCGAGUAAUGGGUAUCUGAUAGCCGAGCUACUCGUCUUCAUCUCUUGCUCGUUCGCAUUUUCCGUGCGGUUCAGCGAGAAGGAGAA ------------..........(((((((.(.(((........))).)..))))))).....((((((.(((((((((((.....))))))....))))).)))))). ( -31.50) >consensus ____________UGCUCAAAUUGCGAGUAAUGGGUAUCUGAUAGUCGAGCUACUCGUCUUCAUCUCUUGCUCGUUCGCAUUUUCCGUGCGGUUCAGCGAGAAGGAGAA ............(((((.......))))).((((((.((........)).))))))(((((...(((((((...((((((.....))))))...))))))).))))). (-19.18 = -21.30 + 2.12)

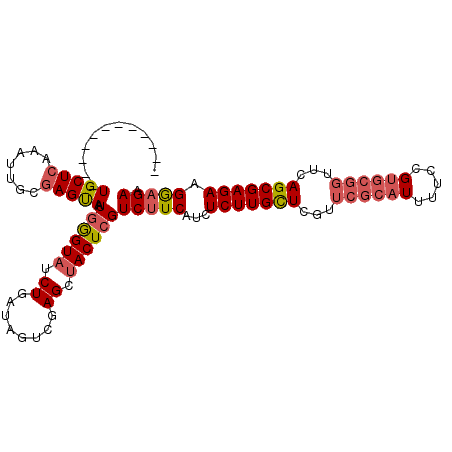
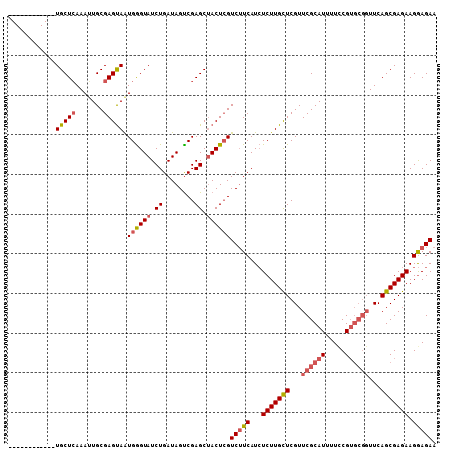
| Location | 5,778,262 – 5,778,373 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.56 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -17.31 |
| Energy contribution | -18.10 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5778262 111 - 22407834 AAAAUUUGGUGUCCAAUUAAGGUAUACUCCUCUAGUGGAACUGUUGCUCAAAGUGCGAGCAAUGAGUAUCUGAUAGUCGAGCCACUAGUC-UCAUAUCUUGCUCGUUCGCAU .......(((.((((....(((......)))....)))))))..........((((((((...(((((...((((...(((.(....).)-)).)))).))))))))))))) ( -28.40) >DroSec_CAF1 15360 95 - 1 A-AAUUUGGUGAGCAAUUAAGAUAUACU----------------UGCUCAAAUUGCGAGUAGUGGGUAUCUGAUAGUCGAGCUACUCGUCUUCAUCUCUUGUUCGUUCGCAU .-......((((((...(((((..((((----------------(((.......))))))).........(((.((.((((...)))).)))))..)))))...)))))).. ( -25.30) >DroSim_CAF1 16129 95 - 1 A-AAUUUGGUGAGCAAUUAAGAUAUACU----------------UGCUAAAAUUGCGAGUAAUGGGUAUCUGAUAGCCGAGCUACUCGUCUUCAUCUCUUGCUCGUUCGCAU .-......((((((..........((((----------------(((.......)))))))..(((((...(((((.((((...)))).))..)))...))))))))))).. ( -27.00) >consensus A_AAUUUGGUGAGCAAUUAAGAUAUACU________________UGCUCAAAUUGCGAGUAAUGGGUAUCUGAUAGUCGAGCUACUCGUCUUCAUCUCUUGCUCGUUCGCAU ........((((((...............................((.......))((((((.(((....(((.((.((((...)))).))))).))))))))))))))).. (-17.31 = -18.10 + 0.79)

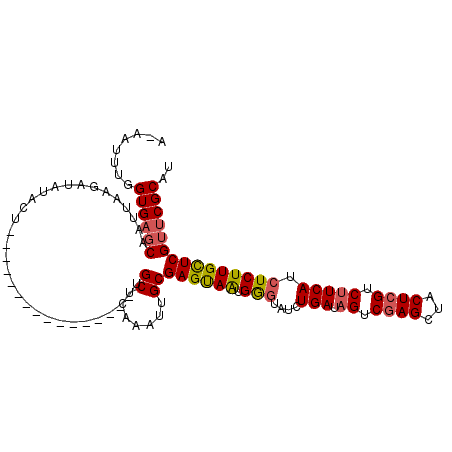
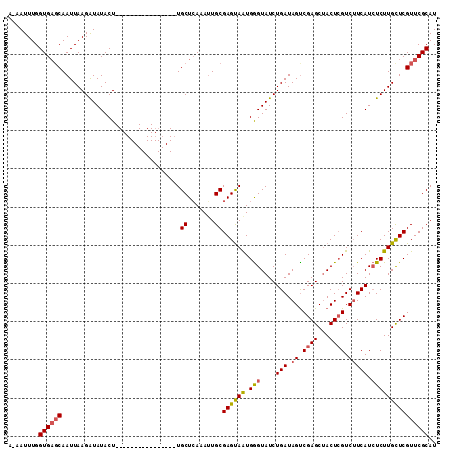
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:36 2006