
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,772,847 – 5,772,957 |
| Length | 110 |
| Max. P | 0.958461 |

| Location | 5,772,847 – 5,772,957 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 84.82 |
| Mean single sequence MFE | -22.06 |
| Consensus MFE | -16.52 |
| Energy contribution | -16.64 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5772847 110 + 22407834 CUGACCUUACACCUGAAUAUCGCAUAUAAAAAUAUUAUUUUCCAGGCACAAAGUAUAUUUUCUUUGCCAAAAAAAAAAAAACAACAAUUGGCAAGGUGUGUUGGCCAGAU (((.((.(((((((..............(((((((.(((((........))))))))))))...((((((.................)))))))))))))..)).))).. ( -22.43) >DroSec_CAF1 10045 92 + 1 CUGACCUUACACCUGAAUAACGCAUAUAAAAAUAUUAUUUUCCAGCCACGAAGUAUAUUUUCUUUGCCAAAACG------------------AAGGUGUGUUGGCCAGAU (((.((.(((((((.......(((....(((((((.(((((........))))))))))))...))).......------------------.)))))))..)).))).. ( -20.56) >DroSim_CAF1 10325 92 + 1 CUGACCUUACACCUGAAUAACGCAUAUAAAAAUAUUAUUUUCCAGGCACGAAGUAUAUUUUCUUUGCCAAAACG------------------AAGGUGUGUUGGCCAGAU (((.((.(((((((((((((..............))))))....((((.((((.....))))..))))......------------------.)))))))..)).))).. ( -20.64) >DroEre_CAF1 10008 107 + 1 CUGACCUUACACCUGAAUACAACAUAUAAAAAUAUUAUUUUCCAGGCACGAAGUAUAUUUUCUUUGCCAAAAAG---AAAACAAGAAAUGCCAAGGUGUGUUGGCCAAAU .((.((.(((((((.........((((....)))).........((((.........((((((((.....))))---)))).......)))).)))))))..)).))... ( -20.59) >DroYak_CAF1 10499 107 + 1 UUGACCUUACACCUGAAUAUCACAUAUAAAAAUAUUAUUUUCCAGGCACGGAGUAUAUUUUCUUUGCCAAAAAG---AACACAACAAAUGCCAAGGUGUGUUGGUCAGAU ((((((.(((((((.(((((...........)))))........((((.((((......)))).))))......---................)))))))..)))))).. ( -26.10) >consensus CUGACCUUACACCUGAAUAACGCAUAUAAAAAUAUUAUUUUCCAGGCACGAAGUAUAUUUUCUUUGCCAAAAAG___AA_ACAA_AA_UG_CAAGGUGUGUUGGCCAGAU (((.((.(((((((.........((((....)))).........((((..(((........))))))).........................)))))))..)).))).. (-16.52 = -16.64 + 0.12)

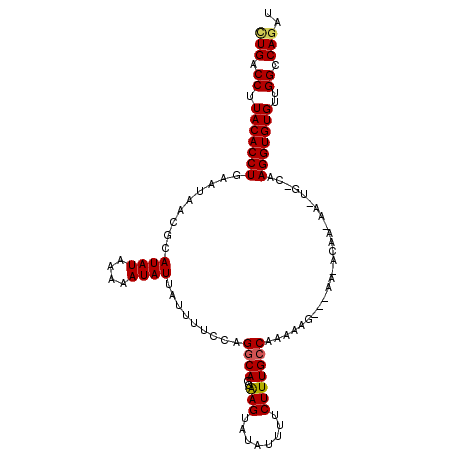
| Location | 5,772,847 – 5,772,957 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 84.82 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -18.92 |
| Energy contribution | -18.88 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5772847 110 - 22407834 AUCUGGCCAACACACCUUGCCAAUUGUUGUUUUUUUUUUUUUGGCAAAGAAAAUAUACUUUGUGCCUGGAAAAUAAUAUUUUUAUAUGCGAUAUUCAGGUGUAAGGUCAG ..((((((......(.(((((((.................))))))).)..........(((..((((((.....((((....))))......))))))..))))))))) ( -27.63) >DroSec_CAF1 10045 92 - 1 AUCUGGCCAACACACCUU------------------CGUUUUGGCAAAGAAAAUAUACUUCGUGGCUGGAAAAUAAUAUUUUUAUAUGCGUUAUUCAGGUGUAAGGUCAG ..((((((...((((((.------------------..(((..((...(((.......)))...))..)))(((.((((....))))..)))....))))))..)))))) ( -22.90) >DroSim_CAF1 10325 92 - 1 AUCUGGCCAACACACCUU------------------CGUUUUGGCAAAGAAAAUAUACUUCGUGCCUGGAAAAUAAUAUUUUUAUAUGCGUUAUUCAGGUGUAAGGUCAG ..((((((((.((.....------------------.)).)))))...........((((..((((((((.(((.((((....))))..))).))))))))..))))))) ( -23.60) >DroEre_CAF1 10008 107 - 1 AUUUGGCCAACACACCUUGGCAUUUCUUGUUUU---CUUUUUGGCAAAGAAAAUAUACUUCGUGCCUGGAAAAUAAUAUUUUUAUAUGUUGUAUUCAGGUGUAAGGUCAG ...(((((...((((((.(((((....((((((---((((.....))))))))))......)))))..(((.(((((((......))))))).)))))))))..))))). ( -32.70) >DroYak_CAF1 10499 107 - 1 AUCUGACCAACACACCUUGGCAUUUGUUGUGUU---CUUUUUGGCAAAGAAAAUAUACUCCGUGCCUGGAAAAUAAUAUUUUUAUAUGUGAUAUUCAGGUGUAAGGUCAA ...(((((...(((((((.((((.((((((.((---((....((((..((........))..)))).)))).)))))).......)))).).....))))))..))))). ( -27.30) >consensus AUCUGGCCAACACACCUUG_CA_UU_UUGU_UU___CUUUUUGGCAAAGAAAAUAUACUUCGUGCCUGGAAAAUAAUAUUUUUAUAUGCGAUAUUCAGGUGUAAGGUCAG ..((((((...(((((..........................((((..((........))..))))((((.....((((....))))......)))))))))..)))))) (-18.92 = -18.88 + -0.04)

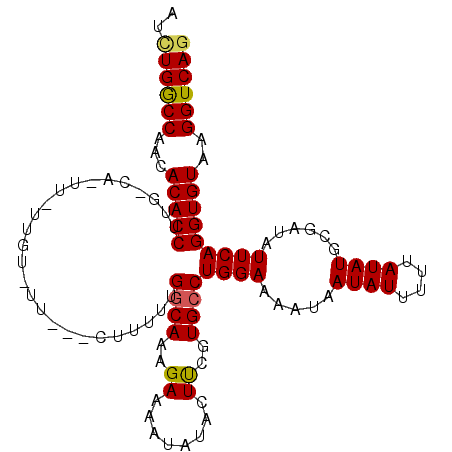

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:30 2006