
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,771,137 – 5,771,390 |
| Length | 253 |
| Max. P | 0.986106 |

| Location | 5,771,137 – 5,771,234 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 80.31 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -20.04 |
| Energy contribution | -20.48 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5771137 97 + 22407834 AUGCUGCAGCGCAACAACCAAUUGCGGCGGGUGAACAUUUUCUGGCCCAGAGGCUCAAAACGCAUGUUGUAGCUGCA----AAAAAA--A-AUUAACAUUAAAA ....((((((((((((......((((..((((.(........).)))).(.....)....)))))))))).))))))----......--.-............. ( -25.40) >DroVir_CAF1 10798 98 + 1 AUGCUGCAGCGCAACAGCCAAUUGCGGCGUGUAAACAUUUUCUGACCCAGCGGCUCAAAGCGCAUGUUGAAGCUGCA----AGCAGA--CAAUUGUUAAUUAAU .(((((((((.((((((((......))).....................(((........))).)))))..))))).----))))((--(....)))....... ( -27.50) >DroPse_CAF1 9912 93 + 1 AUGCUGCAGCGCAGCAGCCAAUUACGGCGCGAGAACAUUUUCUGGCCCAAAGGCUCAAAGCGCAUGUUGUAGCUGCA----AAGAAA--AUGCGGG-----AAA ....(((((((((((((((......)))(((((((....))))((((....)))).....))).)))))).))))))----......--.......-----... ( -30.70) >DroEre_CAF1 8397 100 + 1 AUGCUGCAGCGCAGCAACCAGUUGCGGCGUGUGAACAUUUUCUGGCCCAGAGGCUCGAAACGCAUGUUGUAGCUAUA----AAAAAAAUAAGUUGGCAUUAACG .((((((...))))))...((((((((((((((.....((((.((((....)))).))))))))))))))))))...----..........((((....)))). ( -33.60) >DroYak_CAF1 8524 104 + 1 AUGCUGCAGCGCAGCAACCAGUUGCGGCGUGUGAACAUUUUCUGGCCCAGAGGCUCAAAACGCAUGUUGUAGCUGCAAAGAAAAAAAAAAACAUUGUAAUAACG .((((((...))))))..(((((((((((((((.....(((..((((....))))..))))))))))))))))))............................. ( -32.20) >DroPer_CAF1 9883 93 + 1 AUGCUGCAGCGCAGCAGCCAAUUACGGCGCGAGAACAUUUUCUGGCCCAAAGGCUCAAAGCGCAUGUUGUAGCUGCA----AAGAAA--AUGCGGG-----AAA ....(((((((((((((((......)))(((((((....))))((((....)))).....))).)))))).))))))----......--.......-----... ( -30.70) >consensus AUGCUGCAGCGCAGCAACCAAUUGCGGCGUGUGAACAUUUUCUGGCCCAGAGGCUCAAAACGCAUGUUGUAGCUGCA____AAAAAA__AAGUUGG_A_UAAAA ..((((((((((((.......)))).((((..(((....))).((((....))))....))))..))))))))............................... (-20.04 = -20.48 + 0.44)

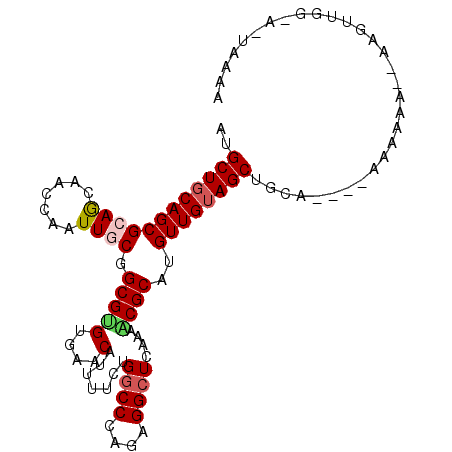

| Location | 5,771,137 – 5,771,234 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.31 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -22.29 |
| Energy contribution | -21.90 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5771137 97 - 22407834 UUUUAAUGUUAAU-U--UUUUUU----UGCAGCUACAACAUGCGUUUUGAGCCUCUGGGCCAGAAAAUGUUCACCCGCCGCAAUUGGUUGUUGCGCUGCAGCAU .............-.--.....(----((((((...(((((...(((((.(((....)))))))).)))))........(((((.....))))))))))))... ( -27.40) >DroVir_CAF1 10798 98 - 1 AUUAAUUAACAAUUG--UCUGCU----UGCAGCUUCAACAUGCGCUUUGAGCCGCUGGGUCAGAAAAUGUUUACACGCCGCAAUUGGCUGUUGCGCUGCAGCAU ...............--..((((----.(((((..(((((.....(((((.((...)).)))))............((((....))))))))).))))))))). ( -27.40) >DroPse_CAF1 9912 93 - 1 UUU-----CCCGCAU--UUUCUU----UGCAGCUACAACAUGCGCUUUGAGCCUUUGGGCCAGAAAAUGUUCUCGCGCCGUAAUUGGCUGCUGCGCUGCAGCAU ...-----...((((--((((..----.((.((........))))..((.(((....)))))))))))))......((((....))))((((((...)))))). ( -28.20) >DroEre_CAF1 8397 100 - 1 CGUUAAUGCCAACUUAUUUUUUU----UAUAGCUACAACAUGCGUUUCGAGCCUCUGGGCCAGAAAAUGUUCACACGCCGCAACUGGUUGCUGCGCUGCAGCAU .......................----....(((..(((((...((((..(((....)))..)))))))))..(((((.((((....)))).))).)).))).. ( -21.30) >DroYak_CAF1 8524 104 - 1 CGUUAUUACAAUGUUUUUUUUUUUCUUUGCAGCUACAACAUGCGUUUUGAGCCUCUGGGCCAGAAAAUGUUCACACGCCGCAACUGGUUGCUGCGCUGCAGCAU ..........................(((((((...(((((...(((((.(((....)))))))).))))).....((.((((....)))).)))))))))... ( -29.70) >DroPer_CAF1 9883 93 - 1 UUU-----CCCGCAU--UUUCUU----UGCAGCUACAACAUGCGCUUUGAGCCUUUGGGCCAGAAAAUGUUCUCGCGCCGUAAUUGGCUGCUGCGCUGCAGCAU ...-----...((((--((((..----.((.((........))))..((.(((....)))))))))))))......((((....))))((((((...)))))). ( -28.20) >consensus UUUUA_U_CCAACUU__UUUUUU____UGCAGCUACAACAUGCGCUUUGAGCCUCUGGGCCAGAAAAUGUUCACACGCCGCAAUUGGCUGCUGCGCUGCAGCAU ...........................((((((...(((((....((((.(((....)))))))..))))).....((.(((......))).)))))))).... (-22.29 = -21.90 + -0.39)

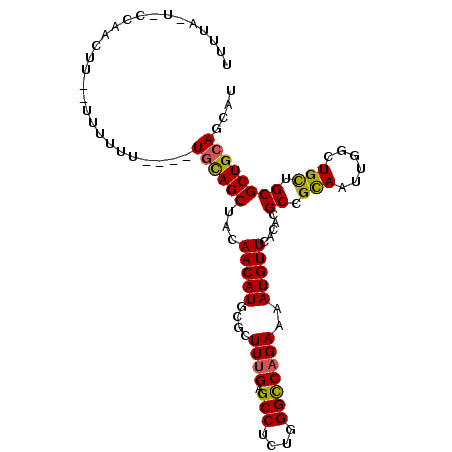

| Location | 5,771,234 – 5,771,351 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.88 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.76 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5771234 117 + 22407834 UAUAUGUUCAUUUGUAAAACAUGAUUU--GAAAAACAACAGAUGCGCGAAACUAAAAUCAUGUCCAUGCUAGCAUGUAGAUAUCCAUGUAGAAAACAUGUAGUUCUACAGCAGCGCCCU ..................(((((((((--.........................)))))))))....(((.((.((((((....(((((.....)))))....)))))))))))..... ( -24.01) >DroSec_CAF1 8348 116 + 1 UAUAUGUCCAA-UGUAAAUCAUAAAUU--GAAUAACAACAGAUGCGCGAAACUAAAAUUAUGUCCAUGCUUGCAUGUAGAUAUCAAUGUAGAAAACAUGUAGUUCUACAGCAGCGCCAU .....(((...-(((.(.(((.....)--)).).)))...)))((((..............((....)).(((.((((((.....((((.....)))).....)))))))))))))... ( -20.40) >DroSim_CAF1 8547 116 + 1 UAUAUGUCCAU-UGUAAAUCAUAAUUU--GAAUAACAACAGACGCGCGAAACUAAAAUCAUUUCCAUGCUUGCAUGUAGAUAUCCAUGUAGAAAACAUGUAGUUCUACAGCAGCGCCCU .....(((..(-(((.(.(((.....)--)).).))))..)))((((((........))...........(((.((((((....(((((.....)))))....)))))))))))))... ( -27.60) >DroEre_CAF1 8497 105 + 1 UACA----------UAAACAA--AUCCUAGAAAAGCAUUAAAUGCGGGAAAAUAAAGUUAUUUACAUGCUAGCAUGUAGAUAUACAUGUACAAAACAUGUAGU-CUACAGCAGCGCCC- ....----------.......--...........((((...))))(((.(((((....)))))....(((.((.(((((((.(((((((.....)))))))))-)))))))))).)))- ( -26.70) >DroYak_CAF1 8628 108 + 1 UAUU----------UAAACAAAAAUUUAAAAAAAACAACAAGUGCGGGAACCUAAAAUCAUUUACAUGCUAGCAUGUAGAUAUACAUGUACAAAACAUGUAGUUUUACAGCAGCGCCC- ..((----------((((......))))))...........((((.(((((........(((((((((....))))))))).(((((((.....))))))))))))......))))..- ( -22.20) >consensus UAUAUGU_CA__UGUAAACCAUAAUUU__GAAAAACAACAGAUGCGCGAAACUAAAAUCAUUUCCAUGCUAGCAUGUAGAUAUCCAUGUAGAAAACAUGUAGUUCUACAGCAGCGCCCU ...................................................................(((.((.((((((....(((((.....)))))....)))))))))))..... (-15.52 = -15.76 + 0.24)

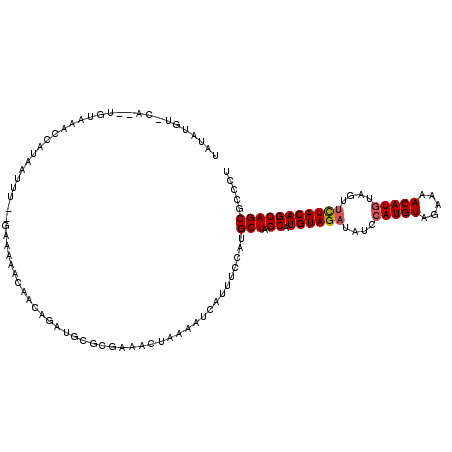

| Location | 5,771,234 – 5,771,351 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.88 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -17.01 |
| Energy contribution | -17.61 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5771234 117 - 22407834 AGGGCGCUGCUGUAGAACUACAUGUUUUCUACAUGGAUAUCUACAUGCUAGCAUGGACAUGAUUUUAGUUUCGCGCAUCUGUUGUUUUUC--AAAUCAUGUUUUACAAAUGAACAUAUA .(..((((((((((((....(((((.....)))))....)))))).)).)))..(((((((((((.((...((.((....))))...)).--))))))))))).......)..)..... ( -28.10) >DroSec_CAF1 8348 116 - 1 AUGGCGCUGCUGUAGAACUACAUGUUUUCUACAUUGAUAUCUACAUGCAAGCAUGGACAUAAUUUUAGUUUCGCGCAUCUGUUGUUAUUC--AAUUUAUGAUUUACA-UUGGACAUAUA (((((((.((((((((.....((((.....)))).....)))))).))((((.((((......)))))))).)))).(((..(((...((--(.....)))...)))-..))))))... ( -22.80) >DroSim_CAF1 8547 116 - 1 AGGGCGCUGCUGUAGAACUACAUGUUUUCUACAUGGAUAUCUACAUGCAAGCAUGGAAAUGAUUUUAGUUUCGCGCGUCUGUUGUUAUUC--AAAUUAUGAUUUACA-AUGGACAUAUA ...(((((((((((((....(((((.....)))))....)))))).)))......(((((.......)))))))))(((((((((...((--(.....)))...)))-))))))..... ( -37.30) >DroEre_CAF1 8497 105 - 1 -GGGCGCUGCUGUAG-ACUACAUGUUUUGUACAUGUAUAUCUACAUGCUAGCAUGUAAAUAACUUUAUUUUCCCGCAUUUAAUGCUUUUCUAGGAU--UUGUUUA----------UGUA -(((.((((((((((-(.(((((((.....)))))))..)))))).)).)))..(((((....)))))...)))((((..(((.((.....)).))--).....)----------))). ( -25.80) >DroYak_CAF1 8628 108 - 1 -GGGCGCUGCUGUAAAACUACAUGUUUUGUACAUGUAUAUCUACAUGCUAGCAUGUAAAUGAUUUUAGGUUCCCGCACUUGUUGUUUUUUUUAAAUUUUUGUUUA----------AAUA -(((.(((..........(((((((.....)))))))(((.((((((....)))))).)))......))).)))...............(((((((....)))))----------)).. ( -22.50) >consensus AGGGCGCUGCUGUAGAACUACAUGUUUUCUACAUGGAUAUCUACAUGCUAGCAUGGAAAUGAUUUUAGUUUCGCGCAUCUGUUGUUUUUC__AAAUUAUGAUUUACA__UG_ACAUAUA .....(((((((((((....(((((.....)))))....)))))).)).))).....((((((((.........(((.....))).......))))))))................... (-17.01 = -17.61 + 0.60)

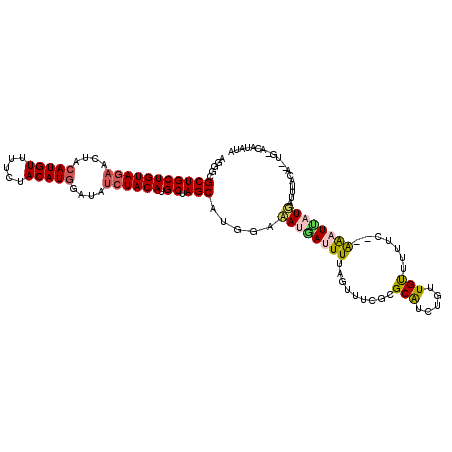

| Location | 5,771,271 – 5,771,390 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.28 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -22.44 |
| Energy contribution | -23.36 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5771271 119 - 22407834 GCCAUCUACCGUAAAGGGUGUUUCUUUUAA-AUCUGUGAGAGGGCGCUGCUGUAGAACUACAUGUUUUCUACAUGGAUAUCUACAUGCUAGCAUGGACAUGAUUUUAGUUUCGCGCAUCU (((.(((((..(((((((....))))))).-....)).))).)))(.(((.(..(((((((((((((.(((((((........)))).)))...)))))))....))))))..)))).). ( -31.20) >DroSec_CAF1 8384 119 - 1 GCCAUCUACCGUAAGGGGUGUUUCUUUUAA-AUCUAUGAGAUGGCGCUGCUGUAGAACUACAUGUUUUCUACAUUGAUAUCUACAUGCAAGCAUGGACAUAAUUUUAGUUUCGCGCAUCU (((((((.((....))(((...........-)))....)))))))(((((((((((.....((((.....)))).....)))))).))).((..((((.........)))).)))).... ( -29.40) >DroSim_CAF1 8583 119 - 1 GCCAUCUACCGUAAGGGGUGUUUCUUUUAA-AUCUAUGAGAGGGCGCUGCUGUAGAACUACAUGUUUUCUACAUGGAUAUCUACAUGCAAGCAUGGAAAUGAUUUUAGUUUCGCGCGUCU ..((((..((....))))))..(((((((.-.....)))))))(((((((((((((....(((((.....)))))....)))))).)))......(((((.......))))))))).... ( -33.30) >DroEre_CAF1 8524 116 - 1 GCCAUCUACAGUACAA-GUGUUUCUUAUAAAAUCUCUGA--GGGCGCUGCUGUAG-ACUACAUGUUUUGUACAUGUAUAUCUACAUGCUAGCAUGUAAAUAACUUUAUUUUCCCGCAUUU ((..((((((((...(-(((((.((((.........)))--))))))))))))))-).(((((((.....)))))))(((.((((((....)))))).))).............)).... ( -30.30) >DroYak_CAF1 8657 118 - 1 GCCAUCUACUGUACAGGGUGUUUCCCAUAAUAUCUCUGA--GGGCGCUGCUGUAAAACUACAUGUUUUGUACAUGUAUAUCUACAUGCUAGCAUGUAAAUGAUUUUAGGUUCCCGCACUU ((...........(((((((((......)))))).))).--(((.(((..........(((((((.....)))))))(((.((((((....)))))).)))......))).))))).... ( -29.40) >consensus GCCAUCUACCGUAAAGGGUGUUUCUUUUAA_AUCUAUGAGAGGGCGCUGCUGUAGAACUACAUGUUUUCUACAUGGAUAUCUACAUGCUAGCAUGGAAAUGAUUUUAGUUUCGCGCAUCU (((.((.....(((((((....)))))))........))...)))(((((((((((....(((((.....)))))....)))))).)).)))............................ (-22.44 = -23.36 + 0.92)

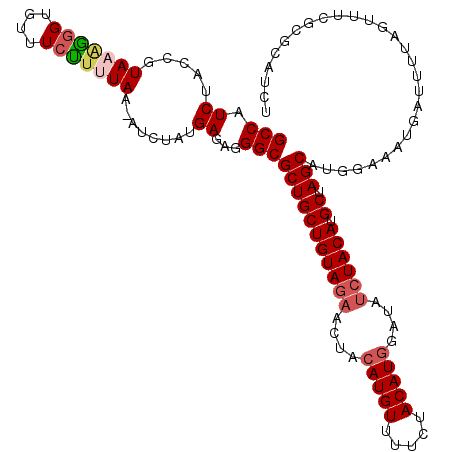
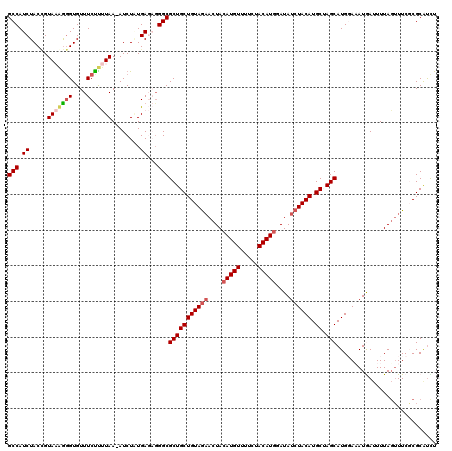
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:28 2006