
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,736,165 – 5,736,271 |
| Length | 106 |
| Max. P | 0.734201 |

| Location | 5,736,165 – 5,736,271 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -20.00 |
| Energy contribution | -19.45 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5736165 106 - 22407834 UGGGACUUUCUCGUGGAGACGACGAGGAAUAGCAAAUUGGCUGUGAUUAUAACCACCCAUUACAUAGAGGAAGCCAAACAGGCAAAUUGUGUAAGUGU---ACAAUGGG ((((...((((((((....).)))))))(((((......)))))...........))))((((((((.....(((.....)))...))))))))....---........ ( -29.40) >DroVir_CAF1 11545 104 - 1 UGGGAAUUUCUGGUGGAAACGACCAGAAAUAGUAAAUUGGCUGUGAUCAUCACCACACAUUAUAUCGAGGAGGCCAAACAGGCCAAUUGCGUGAGUUU---CA--UCUA ..(((((((((((((....).))))))))).(.(((((.((((((........))))..............((((.....))))......)).)))))---).--))). ( -29.00) >DroWil_CAF1 2130 102 - 1 UGGGAUUUUCUUGUGGAAACUACCAGAAAUAGCAAACUGGCUGUGAUUAUUACCACACAUUAUAUUGAGGAAGCCAAGCAGGCUAAUUGUGUAAGUAA---AC-UU--- (((....((((.(((....).)).))))(((((......)))))........))).((.((((((......((((.....))))....))))))))..---..-..--- ( -19.60) >DroMoj_CAF1 13127 105 - 1 UGGGACUUUCUGGUGGAAACGACUAGAAAUAGCAAAUUGGCUGUGAUUAUCACCACACAUUAUAUCGAAGAGGCCAAGCAGGCGAAUUACGUAAGUGG---AA-GUCCA ..(((((((((((((....).))))))....((...((((((.((((................))))....))))))....))...((((....))))---))-)))). ( -30.99) >DroAna_CAF1 7299 108 - 1 UGGAACUUUCUUGUGGAGACCACAAGGAACAGUAAACUGGCUGUGAUCAUCACCACCCACUACAUAGAGGAGGCCAAGCAGGCCAACUGCGUAAGUUGAGGAC-UUGGA (((....(((((((((...)))))))))(((((......)))))........)))((..((......))..))((((((....(((((.....)))))..).)-)))). ( -30.00) >DroPer_CAF1 16622 97 - 1 UGGGAUUUUCUUGUGGAGACAACCAGAAAUAGCAAAUUGGCUGUGAUCAUAACCACACAUUACAUAGAAGAGGCCAAGCAGGCAAAUUGUGUAAG------------GA (((....((((..((....))...))))(((((......)))))........)))....((((((((.....(((.....)))...)))))))).------------.. ( -22.00) >consensus UGGGACUUUCUUGUGGAAACGACCAGAAAUAGCAAAUUGGCUGUGAUCAUCACCACACAUUACAUAGAGGAGGCCAAGCAGGCAAAUUGCGUAAGUGG___AC__U_GA (((....((((((((....).)))))))(((((......)))))........))).................(((.....))).......................... (-20.00 = -19.45 + -0.55)
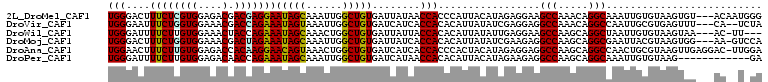
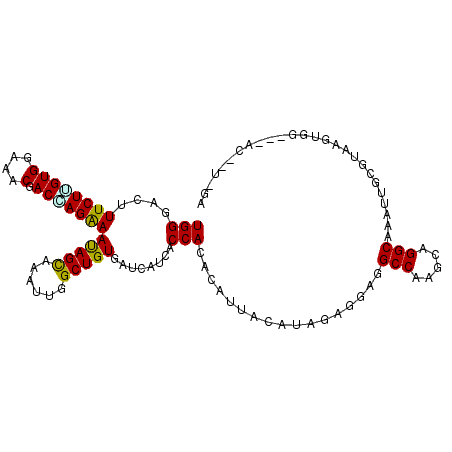
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:19 2006