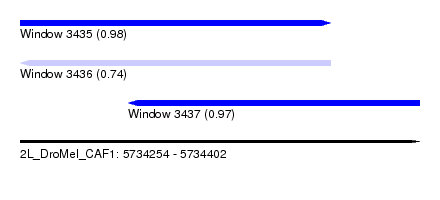
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,734,254 – 5,734,402 |
| Length | 148 |
| Max. P | 0.976746 |

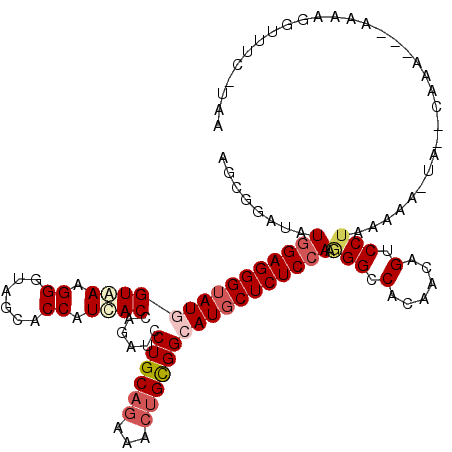
| Location | 5,734,254 – 5,734,369 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.46 |
| Mean single sequence MFE | -34.99 |
| Consensus MFE | -24.45 |
| Energy contribution | -25.45 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
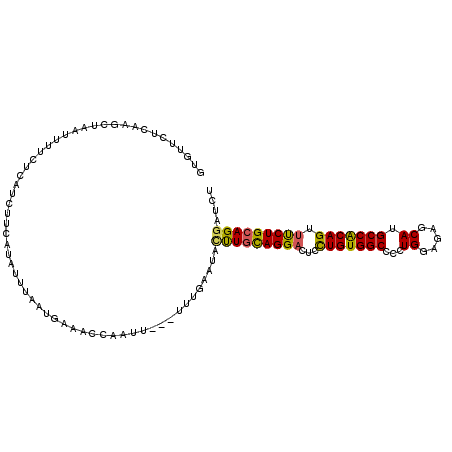
>2L_DroMel_CAF1 5734254 115 + 22407834 AGCGGAGAUGGAGGGAAUGGUGAAGGGUAGCACCAUCACUAGAUCCUGCAGAAACUGCGGCAUGCUCUCCAAAGGCCACAGCAGUCCUGCAAG-AAUUGAAA---AAUAGGUUUCAUAA .((((.(((((((((..((((((.((......)).))))))....((((((...))))))....))))))....((....)).)))))))...-...(((((---......)))))... ( -35.50) >DroVir_CAF1 9197 113 + 1 CGCCGAUAUCGAGGGUAUGGUAAAGGGUAGCACCAUUACCAGAUCCUGCAGAAAGCGCGGCAUGCUCUCCAACGGCCACAGCAGACCUAAACAAAAUGC------AUUAAAAAUCCUUA .((((.....(((((((((((((.((......)).))))).....((((.......)))).))))))))...))))....(((.............)))------.............. ( -29.02) >DroPse_CAF1 14744 113 + 1 UGCAGAUAUGGAGGGUAUUGUAAAGGGUAAUACCAUCACCAGAUCCUGCAGAAACUGUGGCAUGCUCUCCAGGGGCCACAAUAGACCUAAAAA-UA--CAAAUCAAAAUGGUUU---GA (((((((.(((..(((((((.......)))))))....))).)).))))).....((((((...(((....))))))))).............-..--((((((.....)))))---). ( -30.40) >DroYak_CAF1 6363 115 + 1 AGCGGAGAUGGAGGGUAUGGUGAAGGGUAGCACCAUCACUAGGUCCUGCAGGAACUGCGGCAUGCUCUCCAAGGGCCACAGAAGUCCUGCAGG-UAUUCCAA---AUUAGGUUUCAUUA ......(((((((....((((((.((......)).))))))((.(((((((((.(((.(((...((.....)).))).)))...)))))))))-....))..---......))))))). ( -42.10) >DroAna_CAF1 5571 110 + 1 GGCGGAUAUGGAGGGUAUUGUGAAGGGCAGCACCAUCACUAGGUCCUGCAGGAACUGCGGCAUGCUCUCCAGGGGCCACAAGAGUCCUGAAAA-UA-----G---AAAUGGUUCUGUAG .(((((..((((((((((.((((.((......)).))))......((((((...)))))).))))))))))((((((....).))))).....-..-----.---.......))))).. ( -37.20) >DroPer_CAF1 14676 113 + 1 UGCAGAUAUGGAGGGUAUGGUAAAGGGUAAUACCAUCACCAGAUCCUGCAGAAACUGUGGCAUGCUCUCCAGGGGCCACAAUAGGCCUAAAAA-UA--CAAAUCAAAAUUGUUU---GA ..(((((((((((((((((.....((......))...........(..(((...)))..)))))))))))).(((((......))))).....-..--...........)))))---). ( -35.70) >consensus AGCGGAUAUGGAGGGUAUGGUAAAGGGUAGCACCAUCACCAGAUCCUGCAGAAACUGCGGCAUGCUCUCCAAGGGCCACAACAGUCCUAAAAA_UA__CAAA___AAAAGGUUUC_UAA ........(((((((((((((((.((......)).))))......((((((...))))))))))))))))).(((.(......).)))............................... (-24.45 = -25.45 + 1.00)

| Location | 5,734,254 – 5,734,369 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.46 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -19.95 |
| Energy contribution | -18.95 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5734254 115 - 22407834 UUAUGAAACCUAUU---UUUCAAUU-CUUGCAGGACUGCUGUGGCCUUUGGAGAGCAUGCCGCAGUUUCUGCAGGAUCUAGUGAUGGUGCUACCCUUCACCAUUCCCUCCAUCUCCGCU ...(((((......---)))))..(-(((((((((..(((((((((((....)))...))))))))))))))))))....(..((((((........))))))..)............. ( -35.90) >DroVir_CAF1 9197 113 - 1 UAAGGAUUUUUAAU------GCAUUUUGUUUAGGUCUGCUGUGGCCGUUGGAGAGCAUGCCGCGCUUUCUGCAGGAUCUGGUAAUGGUGCUACCCUUUACCAUACCCUCGAUAUCGGCG ............((------((.((..(....((((......))))..)..)).))))((((.((.....))(((...((((((.((......)).))))))...)))......)))). ( -28.90) >DroPse_CAF1 14744 113 - 1 UC---AAACCAUUUUGAUUUG--UA-UUUUUAGGUCUAUUGUGGCCCCUGGAGAGCAUGCCACAGUUUCUGCAGGAUCUGGUGAUGGUAUUACCCUUUACAAUACCCUCCAUAUCUGCA ..---..........((((((--..-....)))))).((((((((..((....))...))))))))....((((.((.(((....((((((.........))))))..))).)))))). ( -25.70) >DroYak_CAF1 6363 115 - 1 UAAUGAAACCUAAU---UUGGAAUA-CCUGCAGGACUUCUGUGGCCCUUGGAGAGCAUGCCGCAGUUCCUGCAGGACCUAGUGAUGGUGCUACCCUUCACCAUACCCUCCAUCUCCGCU ..............---.((((...-(((((((((...(((((((...((.....)).))))))).))))))))).....(((.(((((........))))))))..))))........ ( -40.00) >DroAna_CAF1 5571 110 - 1 CUACAGAACCAUUU---C-----UA-UUUUCAGGACUCUUGUGGCCCCUGGAGAGCAUGCCGCAGUUCCUGCAGGACCUAGUGAUGGUGCUGCCCUUCACAAUACCCUCCAUAUCCGCC ...(((.((((((.---(-----((-(((((((((...(((((((..((....))...))))))).))))).))))..))).)))))).)))........................... ( -27.50) >DroPer_CAF1 14676 113 - 1 UC---AAACAAUUUUGAUUUG--UA-UUUUUAGGCCUAUUGUGGCCCCUGGAGAGCAUGCCACAGUUUCUGCAGGAUCUGGUGAUGGUAUUACCCUUUACCAUACCCUCCAUAUCUGCA ((---(((....)))))..((--((-......((((......))))((((.(((((........).)))).))))....((((.(((((........))))))))).........)))) ( -28.00) >consensus UAA_GAAACCAAUU___UUUG__UA_UUUUCAGGACUACUGUGGCCCCUGGAGAGCAUGCCGCAGUUUCUGCAGGAUCUAGUGAUGGUGCUACCCUUCACCAUACCCUCCAUAUCCGCA ..............................((((...((((((((...((.....)).)))))))).)))).(((.....((((.((......)).)))).....)))........... (-19.95 = -18.95 + -1.00)

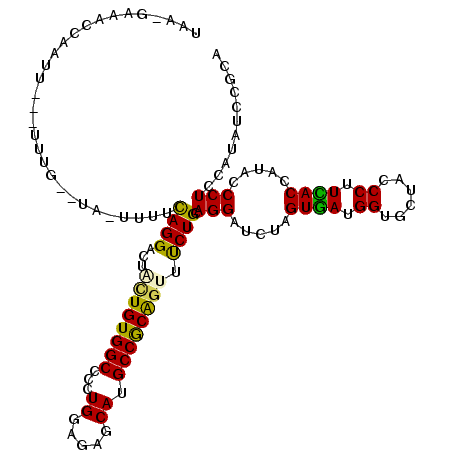
| Location | 5,734,294 – 5,734,402 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.19 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -21.76 |
| Energy contribution | -21.35 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5734294 108 - 22407834 GUGGUGUCAAGCUAAUAUUCUUAUCUUUAUAUUUUAUGAAACCUAUU---UUUCAAUUCUUGCAGGACUGCUGUGGCCUUUGGAGAGCAUGCCGCAGUUUCUGCAGGAUCU ..(((.(((....(((((..........)))))...))).)))....---.......((((((((((..(((((((((((....)))...))))))))))))))))))... ( -31.40) >DroSec_CAF1 10159 108 - 1 GUGUUCUCAAGCUGACUUUCUCAUCUUUAUAUUUUAUGAAACCAAUU---UUUGAAUCAUUGCAGGACUCCUGUGGCCUCUGGAGAGCAUGCCACAGUUUCUGCAGGAUCU ......(((((.(((.....)))..((((((...)))))).......---)))))(((.((((((((...((((((((((....)))...))))))).))))))))))).. ( -25.10) >DroSim_CAF1 5909 108 - 1 GUGUUCUCAAGCUGAUUUUCUCAUCUACAUAUUUCAUUAAACAAAUU---UUUGAAUCCUUGCAGGACUCCUGUGGCCUCUGGAGAGCAUGCCACAGUUUCUGCAGGAUCU ((((.....((.(((.....))).)))))).................---.....(((((.((((((...((((((((((....)))...))))))).))))))))))).. ( -27.80) >DroEre_CAF1 6734 108 - 1 GUGUUUACGAGCCAGUUUACACAUUUUCAUAUUUAAUGAAACCCAAU---UUGGAAUACUUGCAGGACUUCUGUGGCCCCUGGAGAGCAUGCCGCAGUUUCUGCAGGACCU ((((..((......))..))))..((((((.....)))))).((...---..))....(((((((((...(((((((..((....))...))))))).))))))))).... ( -33.30) >DroYak_CAF1 6403 108 - 1 GUGUCCUCCAGCCAGUUUUCCCACAUUCAUAUUUAAUGAAACCUAAU---UUGGAAUACCUGCAGGACUUCUGUGGCCCUUGGAGAGCAUGCCGCAGUUCCUGCAGGACCU ......(((((...((......)).(((((.....))))).......---)))))...(((((((((...(((((((...((.....)).))))))).))))))))).... ( -34.90) >DroPer_CAF1 14716 106 - 1 GUAAAUGUAAUUUCUUUUCCAAAUUAUCACAUUUC---AAACAAUUUUGAUUUG--UAUUUUUAGGCCUAUUGUGGCCCCUGGAGAGCAUGCCACAGUUUCUGCAGGAUCU ......(((((((.......))))))).(((..((---(((....)))))..))--).......((((......))))((((.(((((........).)))).)))).... ( -22.80) >consensus GUGUUCUCAAGCUAAUUUUCUCAUCUUCAUAUUUAAUGAAACCAAUU___UUUGAAUACUUGCAGGACUCCUGUGGCCCCUGGAGAGCAUGCCACAGUUUCUGCAGGAUCU ..........................................................(((((((((...(((((((...((.....)).))))))).))))))))).... (-21.76 = -21.35 + -0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:18 2006