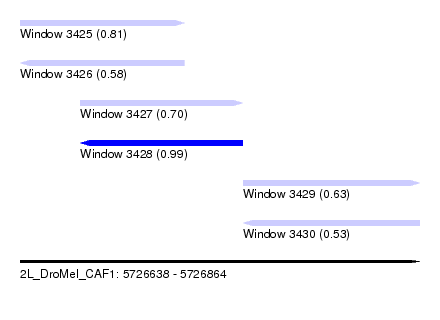
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,726,638 – 5,726,864 |
| Length | 226 |
| Max. P | 0.985707 |

| Location | 5,726,638 – 5,726,731 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -40.52 |
| Consensus MFE | -27.55 |
| Energy contribution | -28.30 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
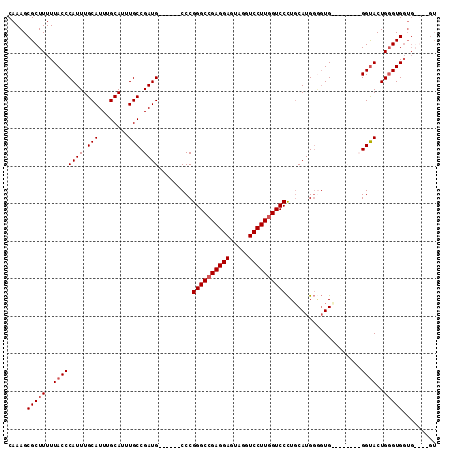
>2L_DroMel_CAF1 5726638 93 + 22407834 CAAAGCGCUUUUUACCCAUUUGCAUUUGCAUUUGCCGAUG------CCCGGGCCGAGGAGUAGGUCCUUGGUCCCUGCAUGGGGUG--------GGCACUGCGUG-------GU ....((((......(((((.(((....)))....((.(((------(..((((((((((.....))))))))))..)))).)))))--------))....)))).-------.. ( -40.10) >DroSim_CAF1 5278 96 + 1 CAAAGCGCUUUUUACCCAUUUGCAUUUGCAUUUGCCGAUG------CCCGGGCCGAGGAGUAGGUCCUUGGUCCCUGCAUGGGGUG--------GGUACUGGGUGGUG----GU .....(((((..(((((((.(((....)))....((.(((------(..((((((((((.....))))))))))..)))).)))))--------))))..)))))...----.. ( -44.00) >DroEre_CAF1 5135 111 + 1 CAAAGCGCUUUUUACCCAUUUGCAUUUGCAUUUGCCGAUGGACGGGCCCGGGCCGAGGAGUAGGUCCUUGGUCCUUCCAUG---UGGAGGUGGUGGUACUGGGUGGUGGGAGGU ......((((((((((((..(((....)))..))(((.....)))((((((((((((((.....))))))))))((((...---.))))...........)))))))))))))) ( -43.10) >DroYak_CAF1 5105 104 + 1 CAAAGCGCUUUUUACCCAUUUGCAUUUGCAUUUGCCGAUG------CCCGGGCAGAGGAGUAGGUCCUUGGUCCCUGCAUGGAGUGUUGUUGGUGGUACUGGGUGGUGGG---- .......((..(((((((..(((....))).(..((((((------((((.((((.(((.(((....))).))))))).))).))...)))))..)...)))))))..))---- ( -34.90) >consensus CAAAGCGCUUUUUACCCAUUUGCAUUUGCAUUUGCCGAUG______CCCGGGCCGAGGAGUAGGUCCUUGGUCCCUGCAUGGGGUG________GGUACUGGGUGGUG____GU .....(((((..((((((((.(((........))).)))).........((((((((((.....))))))))))....................))))..)))))......... (-27.55 = -28.30 + 0.75)

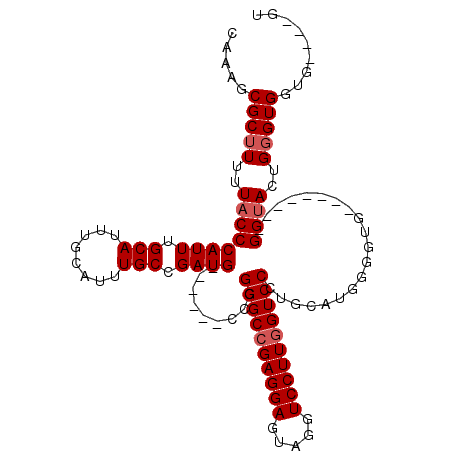
| Location | 5,726,638 – 5,726,731 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.73 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5726638 93 - 22407834 AC-------CACGCAGUGCC--------CACCCCAUGCAGGGACCAAGGACCUACUCCUCGGCCCGGG------CAUCGGCAAAUGCAAAUGCAAAUGGGUAAAAAGCGCUUUG ..-------..(((..((((--------((.((.((((.(((.((.((((.....)))).)))))..)------))).))....(((....)))..))))))....)))..... ( -34.20) >DroSim_CAF1 5278 96 - 1 AC----CACCACCCAGUACC--------CACCCCAUGCAGGGACCAAGGACCUACUCCUCGGCCCGGG------CAUCGGCAAAUGCAAAUGCAAAUGGGUAAAAAGCGCUUUG ..----..........((((--------((.((.((((.(((.((.((((.....)))).)))))..)------))).))....(((....)))..))))))............ ( -30.60) >DroEre_CAF1 5135 111 - 1 ACCUCCCACCACCCAGUACCACCACCUCCA---CAUGGAAGGACCAAGGACCUACUCCUCGGCCCGGGCCCGUCCAUCGGCAAAUGCAAAUGCAAAUGGGUAAAAAGCGCUUUG ..........(((((......((...(((.---...))).((((..((((.....)))).(((....))).))))...))....(((....)))..)))))............. ( -27.30) >DroYak_CAF1 5105 104 - 1 ----CCCACCACCCAGUACCACCAACAACACUCCAUGCAGGGACCAAGGACCUACUCCUCUGCCCGGG------CAUCGGCAAAUGCAAAUGCAAAUGGGUAAAAAGCGCUUUG ----......(((((.................((((((.(((.(..((((.....))))..))))..)------))).))....(((....)))..)))))............. ( -25.00) >consensus AC____CACCACCCAGUACC________CACCCCAUGCAGGGACCAAGGACCUACUCCUCGGCCCGGG______CAUCGGCAAAUGCAAAUGCAAAUGGGUAAAAAGCGCUUUG ..........(((((........................(((.((.((((.....)))).))))).........(((..((....))..)))....)))))............. (-17.98 = -18.73 + 0.75)

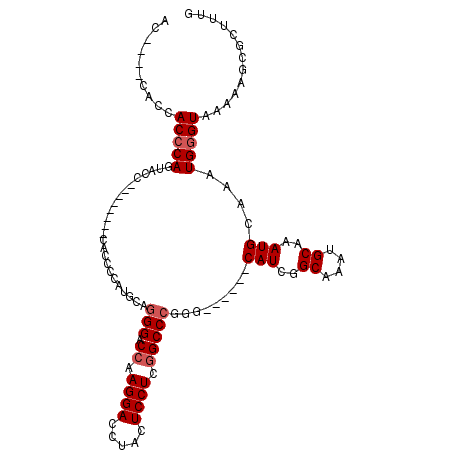
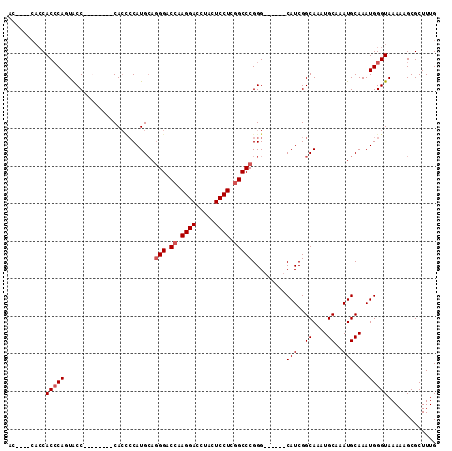
| Location | 5,726,672 – 5,726,764 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -37.73 |
| Consensus MFE | -31.04 |
| Energy contribution | -30.93 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5726672 92 + 22407834 CCGAUGCCCGGGCCGAGGAGUAGGUCCUUGGUCCCUGCAUGGGGUG--------GGCACUGCGUG-----GUGGGCU-AGUGAACAGUGGGCAGUGGGUGGAUUCA ((.((((..((((((((((.....))))))))))..)))).))...--------..((((((.((-----.((..(.-...)..)).)).)))))).......... ( -41.30) >DroSim_CAF1 5312 95 + 1 CCGAUGCCCGGGCCGAGGAGUAGGUCCUUGGUCCCUGCAUGGGGUG--------GGUACUGGGUGGUG--GUGGGCU-AGUGAACAGUGGGCAGUGGGUGUAUUCA (((.(((((((((((((((.....))))))))))(..(.....)..--------)..((((..((.((--(....))-).))..))))))))).)))......... ( -39.40) >DroYak_CAF1 5139 99 + 1 CCGAUGCCCGGGCAGAGGAGUAGGUCCUUGGUCCCUGCAUGGAGUGUUGUUGGUGGUACUGGGUGGUGGG-------CAGUAGGCAGUGGGCAGUGGGUGGAUUCA (((.((((((.((.(((((.....))))).(((((..(....((((..(....)..))))..)..).)))-------).....))..)))))).)))......... ( -32.50) >consensus CCGAUGCCCGGGCCGAGGAGUAGGUCCUUGGUCCCUGCAUGGGGUG________GGUACUGGGUGGUG__GUGGGCU_AGUGAACAGUGGGCAGUGGGUGGAUUCA (((.(((((((((((((((.....))))))))))...(((..((((..........))))..))).......................))))).)))......... (-31.04 = -30.93 + -0.11)



| Location | 5,726,672 – 5,726,764 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -22.87 |
| Energy contribution | -23.20 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5726672 92 - 22407834 UGAAUCCACCCACUGCCCACUGUUCACU-AGCCCAC-----CACGCAGUGCC--------CACCCCAUGCAGGGACCAAGGACCUACUCCUCGGCCCGGGCAUCGG .........((..(((((..((..((((-.((....-----...))))))..--------)).........(((.((.((((.....)))).))))))))))..)) ( -29.00) >DroSim_CAF1 5312 95 - 1 UGAAUACACCCACUGCCCACUGUUCACU-AGCCCAC--CACCACCCAGUACC--------CACCCCAUGCAGGGACCAAGGACCUACUCCUCGGCCCGGGCAUCGG .........((..(((((((((......-.......--.......))))...--------...........(((.((.((((.....)))).))))))))))..)) ( -24.35) >DroYak_CAF1 5139 99 - 1 UGAAUCCACCCACUGCCCACUGCCUACUG-------CCCACCACCCAGUACCACCAACAACACUCCAUGCAGGGACCAAGGACCUACUCCUCUGCCCGGGCAUCGG .........((..(((((......(((((-------.........))))).....................(((.(..((((.....))))..)))))))))..)) ( -22.00) >consensus UGAAUCCACCCACUGCCCACUGUUCACU_AGCCCAC__CACCACCCAGUACC________CACCCCAUGCAGGGACCAAGGACCUACUCCUCGGCCCGGGCAUCGG .........((..(((((((((.......................))))......................(((.((.((((.....)))).))))))))))..)) (-22.87 = -23.20 + 0.33)


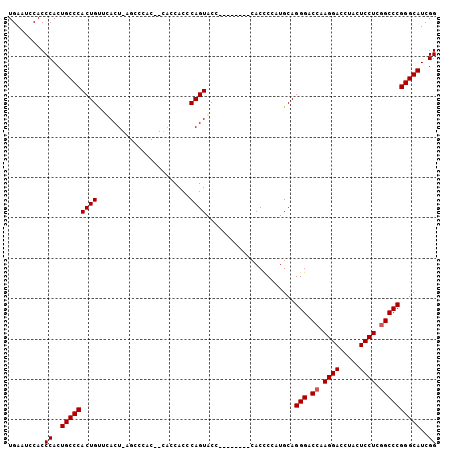
| Location | 5,726,764 – 5,726,864 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.79 |
| Mean single sequence MFE | -35.35 |
| Consensus MFE | -24.69 |
| Energy contribution | -24.50 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5726764 100 + 22407834 UU------------GCGCUUUGGACUGCGUUCAAUAGCCAAAGCGAACGAACAUUUCUGACGUUUGACCGAACUUUGUUGUUGUUCCAGUGCUGCAGCUGCAUCAGCAUCAG-------- ..------------.((((((((.(((.......)))))))))))...........((((.((((....)))).....(((((...((((......))))...)))))))))-------- ( -25.60) >DroSim_CAF1 5407 112 + 1 CUGCGUUUUGCACUGCGCUUUGCACUGCGUUCAAUAGCAAGAGCGAACGAACAUUUCUGACGUUUGACCGAACUUUGUUGUUGUUCCAGUGCUGCAGCUGCAUCAGCAUCAG-------- .(((....(((((((((....((((((.(..((((((((((..((..(((((.........)))))..))...))))))))))..)))))))))))).))))...)))....-------- ( -39.20) >DroEre_CAF1 5286 120 + 1 UUGCGUUUUGCACUGCGCUUUGGACUGCGUUCAAUAGCCAAAGCGCACGAACAUUUCUGACGUUUGACCGAACUUUGUUGUUGUUCCAGUGCUGCAGCUGCAUCAGCAUCAGCAUCAGCA .(((....(((((((((((((((.(((.......))))))))))(((((((((.....((.((((....)))).)).....)))))..)))).)))).))))...)))...((....)). ( -38.70) >DroYak_CAF1 5238 112 + 1 AUGCGUUUUGCACUGCGCUUUGGACUGCGUUCAAUAGCCAAAGCGCACGAACAUUUCUGACGUUUGACCGAACUUUGUUGUUGUUCCAGUGCUGCAGCUGCAUCAGCAUCAG-------- ((((....(((((((((((((((.(((.......))))))))))(((((((((.....((.((((....)))).)).....)))))..)))).)))).))))...))))...-------- ( -37.90) >consensus UUGCGUUUUGCACUGCGCUUUGGACUGCGUUCAAUAGCCAAAGCGAACGAACAUUUCUGACGUUUGACCGAACUUUGUUGUUGUUCCAGUGCUGCAGCUGCAUCAGCAUCAG________ ................((...(.((((.(..(((((((.((((((..(((((.........)))))..))..)))))))))))..))))).).)).((((...))))............. (-24.69 = -24.50 + -0.19)


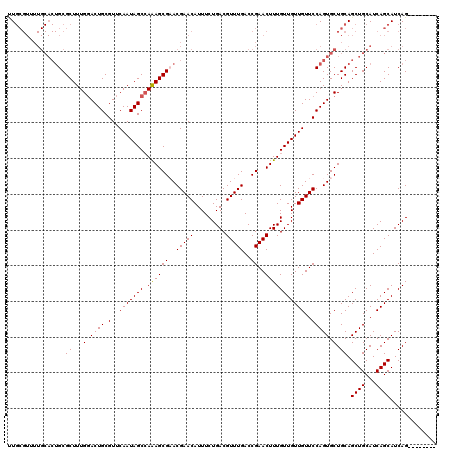
| Location | 5,726,764 – 5,726,864 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.79 |
| Mean single sequence MFE | -32.94 |
| Consensus MFE | -24.23 |
| Energy contribution | -25.72 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5726764 100 - 22407834 --------CUGAUGCUGAUGCAGCUGCAGCACUGGAACAACAACAAAGUUCGGUCAAACGUCAGAAAUGUUCGUUCGCUUUGGCUAUUGAACGCAGUCCAAAGCGC------------AA --------((((((.((((...((....))....((((.........)))).))))..))))))...........((((((((..((((....)))))))))))).------------.. ( -25.10) >DroSim_CAF1 5407 112 - 1 --------CUGAUGCUGAUGCAGCUGCAGCACUGGAACAACAACAAAGUUCGGUCAAACGUCAGAAAUGUUCGUUCGCUCUUGCUAUUGAACGCAGUGCAAAGCGCAGUGCAAAACGCAG --------....(((...(((....)))((((((((((((((........((......)).......)))).))))(((.((((.((((....))))))))))).)))))).....))). ( -31.76) >DroEre_CAF1 5286 120 - 1 UGCUGAUGCUGAUGCUGAUGCAGCUGCAGCACUGGAACAACAACAAAGUUCGGUCAAACGUCAGAAAUGUUCGUGCGCUUUGGCUAUUGAACGCAGUCCAAAGCGCAGUGCAAAACGCAA (((...(((...(((....)))...)))(((((.((((.........))))......(((.((....))..)))(((((((((..((((....)))))))))))))))))).....))). ( -39.50) >DroYak_CAF1 5238 112 - 1 --------CUGAUGCUGAUGCAGCUGCAGCACUGGAACAACAACAAAGUUCGGUCAAACGUCAGAAAUGUUCGUGCGCUUUGGCUAUUGAACGCAGUCCAAAGCGCAGUGCAAAACGCAU --------...((((...(((....)))(((((.((((.........))))......(((.((....))..)))(((((((((..((((....)))))))))))))))))).....)))) ( -35.40) >consensus ________CUGAUGCUGAUGCAGCUGCAGCACUGGAACAACAACAAAGUUCGGUCAAACGUCAGAAAUGUUCGUGCGCUUUGGCUAUUGAACGCAGUCCAAAGCGCAGUGCAAAACGCAA ........((((((.((((...((....))....((((.........)))).))))..)))))).........((((((((((..((((....))))))))))))))............. (-24.23 = -25.72 + 1.50)

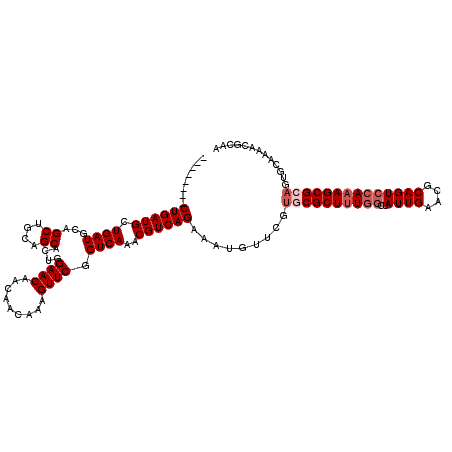
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:11 2006