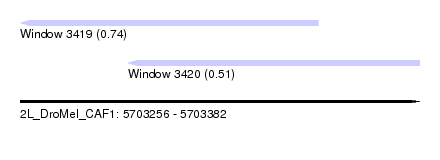
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,703,256 – 5,703,382 |
| Length | 126 |
| Max. P | 0.741117 |

| Location | 5,703,256 – 5,703,350 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 88.59 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -16.26 |
| Energy contribution | -17.90 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5703256 94 - 22407834 CCACGCUAACCACUUUCUGACUUCUUCGCAAGUGAAGUCAGUCAGAAACUGAAAAAUCAG------AUUUUUCCAACGGGUACACACUUGUCCGAAGCUC ....(((......(((((((((.(((((....)))))..)))))))))((((....))))------..........(((..(......)..))).))).. ( -25.10) >DroSec_CAF1 148521 94 - 1 CCACGCUAACCACUUUCUGACUUCUUCGCAAGUGAAGUCAGUCAGAAACUGAAAAAUCUG------AUUUUUCCAACGGGUACACACUGGUCCGAAGCUC ....(((...((.(((((((((.(((((....)))))..))))))))).)).........------.....((..((.(((....))).))..))))).. ( -23.30) >DroSim_CAF1 148143 94 - 1 CCACGCUAACCACUUUCUGACUUCUUCGCAAGUGAAGUCAGUCAGAAACUAAAAAAUCUG------AUUUUUCCAACGGGUACACACUGGUCCGAAGCUC ....(((.((((.(((((((((.(((((....)))))..))))))))).......(((((------..........)))))......))))....))).. ( -22.30) >DroEre_CAF1 151457 100 - 1 UCCCGCUAACCACUUUCUGACUUCUUGGCCAGUAAAGUCAGUCGGAAACUGAAAAAUCUUUGAAACAUUUUUGCAACGGGUACACACUUGUGCAAAGCUC ....(((...((.(((((((((.(((........)))..))))))))).))...................(((((.(((((....))))))))))))).. ( -23.70) >DroYak_CAF1 150123 94 - 1 CCCCGCUAACCAUUUUCUGACUUCCUAGCAAGUGAAGUCAGCCAGAAAGUGAAAAAUCUG------AUUUUUCCAACGGGUACACACUUAUUCGAAGCUU ....(((...((((((((((((((((....)).))))))....)))))))).........------..........((((((......)))))).))).. ( -20.60) >consensus CCACGCUAACCACUUUCUGACUUCUUCGCAAGUGAAGUCAGUCAGAAACUGAAAAAUCUG______AUUUUUCCAACGGGUACACACUUGUCCGAAGCUC ....(((...((.(((((((((.(((((....)))))..))))))))).))....................((..((((((....))))))..))))).. (-16.26 = -17.90 + 1.64)

| Location | 5,703,290 – 5,703,382 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 91.63 |
| Mean single sequence MFE | -20.26 |
| Consensus MFE | -14.89 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5703290 92 - 22407834 UAUUCUAUAUAUUGGUGUUUUUACUUCAGCAGCCACGCUAACCACUUUCUGACUUCUUCGCAAGUGAAGUCAGUCAGAAACUGAAAAAUCAG ............((((((....))...(((......))).)))).(((((((((.(((((....)))))..)))))))))((((....)))) ( -21.60) >DroSec_CAF1 148555 92 - 1 UAUUCUAUAUAUUGGUGUUUUUACUUCAGGAGCCACGCUAACCACUUUCUGACUUCUUCGCAAGUGAAGUCAGUCAGAAACUGAAAAAUCUG ...........(((((((.....(....).....))))))).((.(((((((((.(((((....)))))..))))))))).))......... ( -21.50) >DroSim_CAF1 148177 92 - 1 UAUUCUAUAUAUUGGUGUUUUUACUUCAGGAGCCACGCUAACCACUUUCUGACUUCUUCGCAAGUGAAGUCAGUCAGAAACUAAAAAAUCUG ...........(((((((.....(....).....)))))))....(((((((((.(((((....)))))..)))))))))............ ( -19.80) >DroEre_CAF1 151497 92 - 1 UAUCCUAUACAUUGGUGUUUUUACUUCACGAGUCCCGCUAACCACUUUCUGACUUCUUGGCCAGUAAAGUCAGUCGGAAACUGAAAAAUCUU ...........((((((.....((((...))))..)))))).((.(((((((((.(((........)))..))))))))).))......... ( -15.70) >DroYak_CAF1 150157 92 - 1 UAUUCUAUAUAUUGGUGUUUUCACUUCAGGAGCCCCGCUAACCAUUUUCUGACUUCCUAGCAAGUGAAGUCAGCCAGAAAGUGAAAAAUCUG .............(((.((((((((((.(((((...))).........((((((((((....)).)))))))))).).)))))))))))).. ( -22.70) >consensus UAUUCUAUAUAUUGGUGUUUUUACUUCAGGAGCCACGCUAACCACUUUCUGACUUCUUCGCAAGUGAAGUCAGUCAGAAACUGAAAAAUCUG ...........((((((..................)))))).((.(((((((((.(((((....)))))..))))))))).))......... (-14.89 = -15.73 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:02 2006