
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,693,783 – 5,693,922 |
| Length | 139 |
| Max. P | 0.998795 |

| Location | 5,693,783 – 5,693,893 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 96.91 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -27.10 |
| Energy contribution | -27.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5693783 110 + 22407834 CAUGUUUAUUAAUUUGCUACUAUUUCUGCGCAUUUUUCACAGCUUCUGUUUUCAGCACCUUGGGGGAUUCCCCAUGGCGCACAUCAAAGCAAAUGCCAAAGCGUUGCCAA .((((((....(((((((........(((((..........(((.........)))..(.(((((....))))).))))))......)))))))....))))))...... ( -26.94) >DroSec_CAF1 139012 110 + 1 CAUGUUUAUAAAUUUGCUACUAUUUCUGCGCAAUUUUCAUAGCUUCUGUUUUCAGCACCUUGGGGGAUUCCCCAUGGCGCACAUCAAAGCAAAUGCCAAAGCGUUGCCAA .((((((....(((((((........(((((..........(((.........)))..(.(((((....))))).))))))......)))))))....))))))...... ( -26.94) >DroSim_CAF1 138508 110 + 1 CAUGUUUAUAAAUUUGCUACUAUUUCUGCGCAUUUUUCAUAGCUUCUGUUUUCAGCACCUUGGGGGAUUCCCCACGGCGCACAUCAAAGCAAAUGCCAAAGCGUUGCCAA .((((((....(((((((........(((((..........(((.........)))..(.(((((....))))).))))))......)))))))....))))))...... ( -26.94) >DroEre_CAF1 141652 110 + 1 CAUGUUUAUUAAUUUGCUACUAUUUCUGCGCAUUUUUCACAGCUUCUGUUUUCAGCACCUUGGGGGAUUCCCCAUGGCGCACAUCAAAGCAAAUGCCGAAGCGUGGCCAA (((((((....(((((((........(((((..........(((.........)))..(.(((((....))))).))))))......)))))))....)))))))..... ( -31.54) >DroYak_CAF1 140633 110 + 1 CAUGUUUAUUAAUUUGCUACUAUUUCUGCGCAUUUUUCACAGCUUCUGUUUUCAGCACCUUGGGGGAUUCCCCACCGCGCACAUCAAAGCAAAUGCCAAAGCGUUGCCAA .((((((....(((((((........(((((..........(((.........)))....(((((....)))))..)))))......)))))))....))))))...... ( -26.94) >consensus CAUGUUUAUUAAUUUGCUACUAUUUCUGCGCAUUUUUCACAGCUUCUGUUUUCAGCACCUUGGGGGAUUCCCCAUGGCGCACAUCAAAGCAAAUGCCAAAGCGUUGCCAA .((((((....(((((((........(((((..........(((.........)))....(((((....)))))..)))))......)))))))....))))))...... (-27.10 = -27.10 + -0.00)

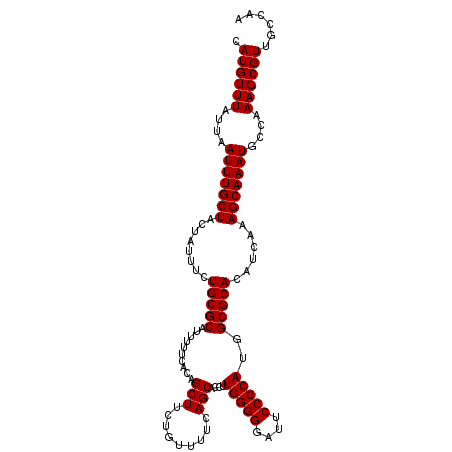
| Location | 5,693,783 – 5,693,893 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 96.91 |
| Mean single sequence MFE | -32.29 |
| Consensus MFE | -30.20 |
| Energy contribution | -30.80 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5693783 110 - 22407834 UUGGCAACGCUUUGGCAUUUGCUUUGAUGUGCGCCAUGGGGAAUCCCCCAAGGUGCUGAAAACAGAAGCUGUGAAAAAUGCGCAGAAAUAGUAGCAAAUUAAUAAACAUG ...((.((..(((((((((((((((.....(((((.(((((....))))).)))))........)))))......)))))).))))....)).))............... ( -32.52) >DroSec_CAF1 139012 110 - 1 UUGGCAACGCUUUGGCAUUUGCUUUGAUGUGCGCCAUGGGGAAUCCCCCAAGGUGCUGAAAACAGAAGCUAUGAAAAUUGCGCAGAAAUAGUAGCAAAUUUAUAAACAUG ..(....)(((((.(((((......)))))(((((.(((((....))))).)))))........)))))((((((..(((((((....).)).)))).))))))...... ( -32.10) >DroSim_CAF1 138508 110 - 1 UUGGCAACGCUUUGGCAUUUGCUUUGAUGUGCGCCGUGGGGAAUCCCCCAAGGUGCUGAAAACAGAAGCUAUGAAAAAUGCGCAGAAAUAGUAGCAAAUUUAUAAACAUG ...((.((..(((((((((((((((.....(((((.(((((....))))).)))))........)))))......)))))).))))....)).))............... ( -32.52) >DroEre_CAF1 141652 110 - 1 UUGGCCACGCUUCGGCAUUUGCUUUGAUGUGCGCCAUGGGGAAUCCCCCAAGGUGCUGAAAACAGAAGCUGUGAAAAAUGCGCAGAAAUAGUAGCAAAUUAAUAAACAUG ...((.(((((((.(((((......)))))(((((.(((((....))))).)))))........)))))((((.......))))......)).))............... ( -34.50) >DroYak_CAF1 140633 110 - 1 UUGGCAACGCUUUGGCAUUUGCUUUGAUGUGCGCGGUGGGGAAUCCCCCAAGGUGCUGAAAACAGAAGCUGUGAAAAAUGCGCAGAAAUAGUAGCAAAUUAAUAAACAUG ...((.((..(((((((((((((((.....((((..(((((....)))))..))))........)))))......)))))).))))....)).))............... ( -29.82) >consensus UUGGCAACGCUUUGGCAUUUGCUUUGAUGUGCGCCAUGGGGAAUCCCCCAAGGUGCUGAAAACAGAAGCUGUGAAAAAUGCGCAGAAAUAGUAGCAAAUUAAUAAACAUG ...((.((..(((((((((((((((.....(((((.(((((....))))).)))))........)))))......)))))).))))....)).))............... (-30.20 = -30.80 + 0.60)

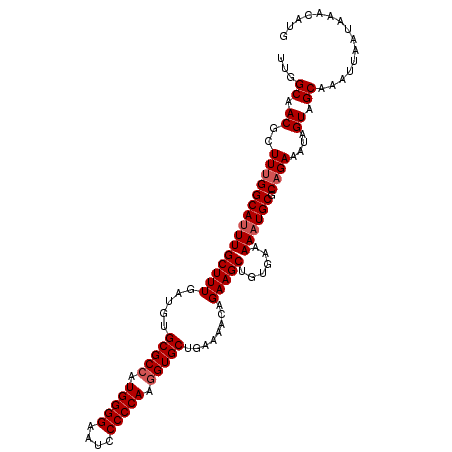

| Location | 5,693,815 – 5,693,922 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -34.91 |
| Consensus MFE | -32.96 |
| Energy contribution | -32.76 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5693815 107 - 22407834 GGACCUGCGGAAAUGGGAACCCUGCGAUGUUGGCAACGCUUUGGCAUUUGCUUUGAUGUGCGCCAUGGGGAAUCCCCCAAGGUGCUGAAAACAGAAGCUGUGAAAAA .....(((.(((..((....)).(((.((....)).)))))).)))...(((((.....(((((.(((((....))))).)))))........)))))......... ( -34.22) >DroSec_CAF1 139044 107 - 1 GGACCUGCAGAAAUGGGAACCCCGCGAUGUUGGCAACGCUUUGGCAUUUGCUUUGAUGUGCGCCAUGGGGAAUCCCCCAAGGUGCUGAAAACAGAAGCUAUGAAAAU .(.((.(((....((((...))))...))).)))...(((((.(((((......)))))(((((.(((((....))))).)))))........)))))......... ( -34.10) >DroSim_CAF1 138540 107 - 1 GGACCUGCAGAAAUGGGAACCCCGCGAUGUUGGCAACGCUUUGGCAUUUGCUUUGAUGUGCGCCGUGGGGAAUCCCCCAAGGUGCUGAAAACAGAAGCUAUGAAAAA .(.((.(((....((((...))))...))).)))...(((((.(((((......)))))(((((.(((((....))))).)))))........)))))......... ( -34.10) >DroEre_CAF1 141684 107 - 1 GGACCUGCAGAAAUGGGAACCCUGCGUUGUUGGCCACGCUUCGGCAUUUGCUUUGAUGUGCGCCAUGGGGAAUCCCCCAAGGUGCUGAAAACAGAAGCUGUGAAAAA ((.((.((((....((....))....)))).))))..(((((.(((((......)))))(((((.(((((....))))).)))))........)))))......... ( -40.60) >DroYak_CAF1 140665 107 - 1 GGACCUGCGGAAAUGGGAACCCUGCGAUGUUGGCAACGCUUUGGCAUUUGCUUUGAUGUGCGCGGUGGGGAAUCCCCCAAGGUGCUGAAAACAGAAGCUGUGAAAAA .....(((.(((..((....)).(((.((....)).)))))).)))...(((((.....((((..(((((....)))))..))))........)))))......... ( -31.52) >consensus GGACCUGCAGAAAUGGGAACCCUGCGAUGUUGGCAACGCUUUGGCAUUUGCUUUGAUGUGCGCCAUGGGGAAUCCCCCAAGGUGCUGAAAACAGAAGCUGUGAAAAA .(.((.(((.....((....)).....))).)))...(((((.(((((......)))))(((((.(((((....))))).)))))........)))))......... (-32.96 = -32.76 + -0.20)

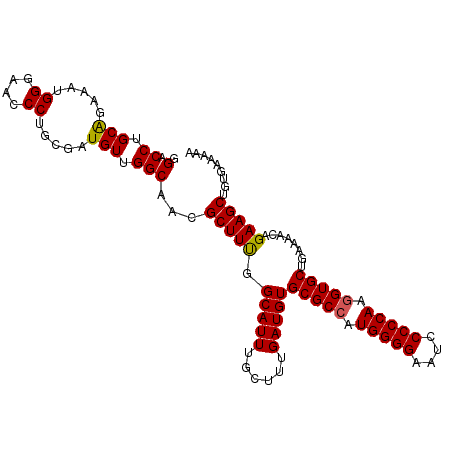
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:55 2006