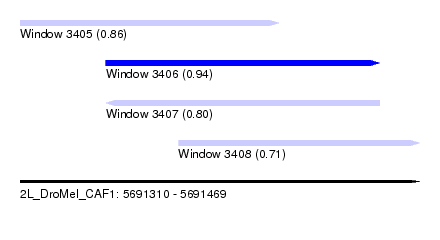
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,691,310 – 5,691,469 |
| Length | 159 |
| Max. P | 0.943082 |

| Location | 5,691,310 – 5,691,413 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.29 |
| Mean single sequence MFE | -37.34 |
| Consensus MFE | -23.82 |
| Energy contribution | -25.06 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5691310 103 + 22407834 UGAUUAAGGCCAGGCGGGUGGAAGUGAGCCCCUG--------CUCUGGAGCUCCGGUGCUGUGGACU---------CCAUGGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCCC ............((((((((((.(.((((.((..--------....)).)))))(((.((((((...---------)))))))))..))))))))))....................... ( -36.10) >DroSec_CAF1 136555 103 + 1 UGAUUAAGGCCAGGCGGGUGGAAGUGAGUCCCUG--------CUCUGGCGCUCCGGAGCUCUGGACU---------CCAUGGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCCC ............((((((((((.((((((((..(--------((((((....)))))))...)))))---------).....))...))))))))))....................... ( -38.60) >DroSim_CAF1 136038 103 + 1 UGAUUAAGGCCAGGCGGGUGGAAGUGAGCCCCUG--------CUCUGGUGCUCCGGAGCUCUGGACU---------CCAUGGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCCC ............((((((((((.(((((((...(--------((((((....)))))))...)).))---------).....))...))))))))))....................... ( -34.90) >DroEre_CAF1 139258 97 + 1 UGAUUAAAGCCAGGCGGA-----------------------GCUCCGGAGCUCCGGAGCUCUGGACUCCAUGGACUCCAUGGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCGC ............((((((-----------------------(((((((....))))))))))(((((((((((...)))))))...))))....)))....................... ( -40.70) >DroYak_CAF1 138138 113 + 1 UGAUUAAGGCCAGGCGGAUGGAAGAGAGCCCCUGCUCCAAAGCUCCAAAGCUCCAGAGCUGUGGACUG-------UCCAUGGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCGC ............((((((((((.(.((((....)))))..((.((((.((((....)))).))))))(-------((....)))...))))))))))....................... ( -36.40) >consensus UGAUUAAGGCCAGGCGGGUGGAAGUGAGCCCCUG________CUCUGGAGCUCCGGAGCUCUGGACU_________CCAUGGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCCC ............((((((((((....................((((((....))))))(..(((............)))..).....))))))))))....................... (-23.82 = -25.06 + 1.24)

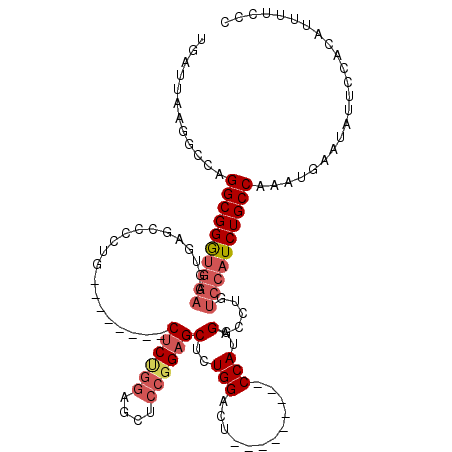
| Location | 5,691,344 – 5,691,453 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.55 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.54 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5691344 109 + 22407834 --CUCUGGAGCUCCGGUGCUGUGGACU---------CCAUGGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCCCAUUCCUUCCGCACCCGCAUAUUUUUCUAUGCGCAAUCACA --....((....))(((((((((((..---------...((((....))))((........))....))))))................)))))((((((......))))))........ ( -24.79) >DroSec_CAF1 136589 109 + 1 --CUCUGGCGCUCCGGAGCUCUGGACU---------CCAUGGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCCCAUUCCUUCCGCACCCGCAUAUUUUUCUAUGCGCAAUCACA --...((((..(((((....)))))..---------..(((((....)))))..))))....................................((((((......))))))........ ( -22.70) >DroSim_CAF1 136072 109 + 1 --CUCUGGUGCUCCGGAGCUCUGGACU---------CCAUGGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCCCAUUCCUUCCGCACCCGCAUAUUUUUCUAUGCGCAAUCACA --....((((((((((....)))))..---------..(((((....))))).....................................)))))((((((......))))))........ ( -25.10) >DroEre_CAF1 139276 119 + 1 -GCUCCGGAGCUCCGGAGCUCUGGACUCCAUGGACUCCAUGGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCGCAAUCCCUCCGCACCCGCAUAUUUUUCUAUGCGCAAUCACA -(((((((....)))))))..((((((((((((...)))))))...)))))..(((.(((((.........)))))..))).............((((((......))))))........ ( -38.50) >DroYak_CAF1 138178 113 + 1 AGCUCCAAAGCUCCAGAGCUGUGGACUG-------UCCAUGGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCGCUAUCCCUCCGCACCCGCAUAUUUUUCUAUGCGCAAUCACA .((.....((((....))))((((((.(-------((....)))..))))))..((.(((((.........)))))..)).........))...((((((......))))))........ ( -24.80) >consensus __CUCUGGAGCUCCGGAGCUCUGGACU_________CCAUGGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCCCAUUCCUUCCGCACCCGCAUAUUUUUCUAUGCGCAAUCACA ...(((((((((....))))))))).............(((((....))))).(((.(((((.........)))))...................(((((......))))))))...... (-19.94 = -20.54 + 0.60)

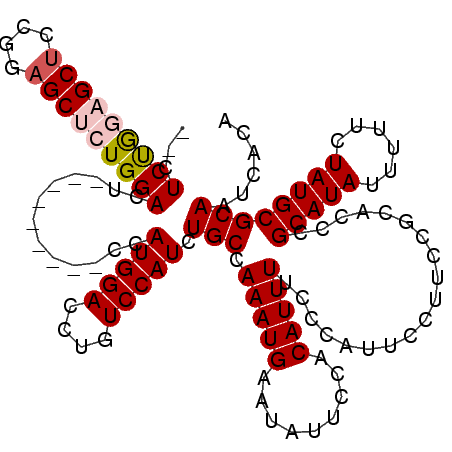

| Location | 5,691,344 – 5,691,453 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.55 |
| Mean single sequence MFE | -35.31 |
| Consensus MFE | -25.64 |
| Energy contribution | -25.16 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5691344 109 - 22407834 UGUGAUUGCGCAUAGAAAAAUAUGCGGGUGCGGAAGGAAUGGGAAAAUGUGGAAUAUUCAUUUGGCAGAUGGACAGGUCCAUGG---------AGUCCACAGCACCGGAGCUCCAGAG-- ........((((((......))))))((.((....((..(((((...((((((...((((((.....))))))....)))))).---------..))).))...))...)).))....-- ( -31.40) >DroSec_CAF1 136589 109 - 1 UGUGAUUGCGCAUAGAAAAAUAUGCGGGUGCGGAAGGAAUGGGAAAAUGUGGAAUAUUCAUUUGGCAGAUGGACAGGUCCAUGG---------AGUCCAGAGCUCCGGAGCGCCAGAG-- ........((((((......))))))(((((....(((.(.(((...((((((...((((((.....))))))....)))))).---------..)))...).)))...)))))....-- ( -34.80) >DroSim_CAF1 136072 109 - 1 UGUGAUUGCGCAUAGAAAAAUAUGCGGGUGCGGAAGGAAUGGGAAAAUGUGGAAUAUUCAUUUGGCAGAUGGACAGGUCCAUGG---------AGUCCAGAGCUCCGGAGCACCAGAG-- ........((((((......))))))(((((....(((.(.(((...((((((...((((((.....))))))....)))))).---------..)))...).)))...)))))....-- ( -35.20) >DroEre_CAF1 139276 119 - 1 UGUGAUUGCGCAUAGAAAAAUAUGCGGGUGCGGAGGGAUUGCGAAAAUGUGGAAUAUUCAUUUGGCAGAUGGACAGGUCCAUGGAGUCCAUGGAGUCCAGAGCUCCGGAGCUCCGGAGC- .(..(((.((((((......)))))(....)...).)))..).....(((.((((....)))).)))..(((((...(((((((...))))))))))))..(((((((....)))))))- ( -42.40) >DroYak_CAF1 138178 113 - 1 UGUGAUUGCGCAUAGAAAAAUAUGCGGGUGCGGAGGGAUAGCGAAAAUGUGGAAUAUUCAUUUGGCAGAUGGACAGGUCCAUGGA-------CAGUCCACAGCUCUGGAGCUUUGGAGCU ........((((((......))))))..(((.((((((((..............))))).))).)))..(((((..(((....))-------).))))).(((((..(....)..))))) ( -32.74) >consensus UGUGAUUGCGCAUAGAAAAAUAUGCGGGUGCGGAAGGAAUGGGAAAAUGUGGAAUAUUCAUUUGGCAGAUGGACAGGUCCAUGG_________AGUCCAGAGCUCCGGAGCUCCAGAG__ ........((((((......))))))((.((....(((.........(((.((((....)))).))).(((((....))))).............)))...)).))((....))...... (-25.64 = -25.16 + -0.48)

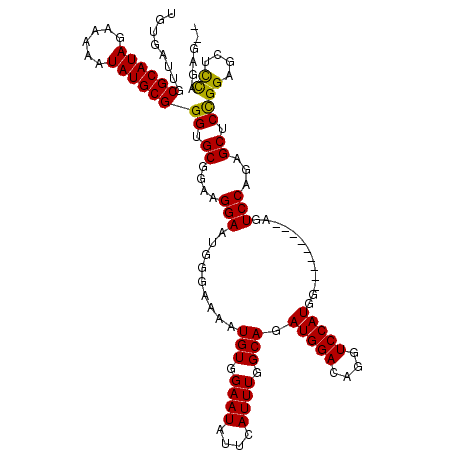

| Location | 5,691,373 – 5,691,469 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -18.16 |
| Consensus MFE | -16.64 |
| Energy contribution | -16.64 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5691373 96 + 22407834 GGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCCCAUUCCUUCCGCACCCGCAUAUUUUUCUAUGCGCAAUCACAACAGAUUGUGUGUUGG (((....))).....................................((((((..(((((......)))))((((((......))))))))).))) ( -17.70) >DroSec_CAF1 136618 96 + 1 GGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCCCAUUCCUUCCGCACCCGCAUAUUUUUCUAUGCGCAAUCACAACAGAUUGUGUGUUGG (((....))).....................................((((((..(((((......)))))((((((......))))))))).))) ( -17.70) >DroSim_CAF1 136101 96 + 1 GGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCCCAUUCCUUCCGCACCCGCAUAUUUUUCUAUGCGCAAUCACAACAGAUUGUGUGUUGG (((....))).....................................((((((..(((((......)))))((((((......))))))))).))) ( -17.70) >DroEre_CAF1 139315 96 + 1 GGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCGCAAUCCCUCCGCACCCGCAUAUUUUUCUAUGCGCAAUCACAACAGAUUGUGUGUUGC (((....)))...(((.(((((.........)))))..)))........((((..(((((......)))))((((((......))))))))))... ( -19.50) >DroYak_CAF1 138211 96 + 1 GGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCGCUAUCCCUCCGCACCCGCAUAUUUUUCUAUGCGCAAUCACAACAGAUUGUGUGUUGC (((....)))....((.(((((.........)))))..)).........((((..(((((......)))))((((((......))))))))))... ( -18.20) >consensus GGACCUGUCCAUCUGCCAAAUGAAUAUUCCACAUUUUCCCAUUCCUUCCGCACCCGCAUAUUUUUCUAUGCGCAAUCACAACAGAUUGUGUGUUGG (((....))).....................................(.((((..(((((......)))))((((((......)))))))))).). (-16.64 = -16.64 + 0.00)

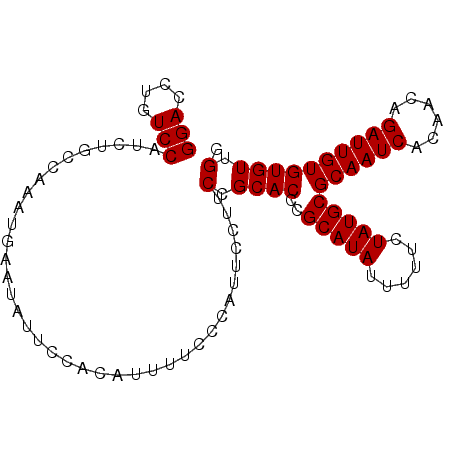
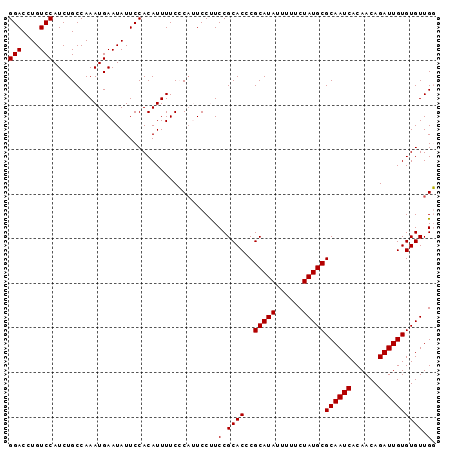
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:50 2006