| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,641,985 – 5,642,086 |
| Length | 101 |
| Max. P | 0.984117 |

| Location | 5,641,985 – 5,642,086 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.24 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -15.37 |
| Energy contribution | -14.76 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5641985 101 + 22407834 UUUCUUAGUCUAAUCUAAAACAAGGUAAAUAUCAGGUUGUUUCCUGUAUUCG---AUCGAAACAUCUGUAUAUACCUUUGUUCCGAUU-GGACAAAA--GUGCGUC-U .......((((((((...(((((((((.(((.(((((.(((((.........---...))))))))))))).)))).)))))..))))-))))....--.......-. ( -27.30) >DroVir_CAF1 125989 106 + 1 AACAACAAUCUAAUCUGAAACAAGGUACAUAUCAGAGUGUUUCUUAAAUUUGAUUAUUGAAACCUCUGUAUAUACCUUUCAUUGGAUU-AAACUGAUAAGCACAUG-U ..........(((((..(...((((((.(((.(((((.(((((.(((......)))..))))))))))))).))))))...)..))))-)................-. ( -24.80) >DroEre_CAF1 88237 104 + 1 GUUCUUAGUCUAAUCUAAAACAAGGUACAUAUCAGGUUGUUUCCUGUAUUGGAUGAUCGAAACAUCUGUAUAUACCUUUGUUCCGAAU-GGACGAAA--GAGCAUC-U ((((((.(((((.((...(((((((((.(((.(((((.(((((...(((...)))...))))))))))))).)))).)))))..)).)-))))..))--))))...-. ( -30.20) >DroWil_CAF1 126026 106 + 1 AUUCUAAUUCUAGUCUAAAACAAGGUACAUAUCAGGUUGUUUCGUAUAUUUGAAGCCUGAAACAUCUGUAUCUACCUUUGUUUCGAAUAGGACUAAA--GAGUUUUCU .....(((((((((((.((((((((((.(((.(((((.((((((.............)))))))))))))).)))).))))))......)))))..)--))))).... ( -24.02) >DroYak_CAF1 87666 104 + 1 GUUCUUGGUCUAAUCUAAAACAAGGUACAUAUCAGGUUGUUUCCUAUAUUGGAUGAUCGAAACAUCUGUAUAUACCUUUGUUCCGAAU-GGACGAAA--GAGCAUC-U ((((((.(((((.((...(((((((((.(((.(((((.(((((...............))))))))))))).)))).)))))..)).)-))))..))--))))...-. ( -30.06) >DroAna_CAF1 83618 104 + 1 GUUCUUGAUUUAAUCUAAAACAAGGUACAUAUCUGGUUGUUUCCUAUAUUCAAAGUUUGAAACAUCUGUAUCUACCUUUGUUUCGAUU-AAACUAAA--GAGCUAC-U ((((((..(((((((..((((((((((.((((..((.((((((..((.......))..)))))).)))))).)))).)))))).))))-)))...))--))))...-. ( -26.10) >consensus GUUCUUAGUCUAAUCUAAAACAAGGUACAUAUCAGGUUGUUUCCUAUAUUCGAUGAUCGAAACAUCUGUAUAUACCUUUGUUCCGAAU_GGACUAAA__GAGCAUC_U .....................((((((.(((.(((((.(((((...............))))))))))))).)))))).((((......))))............... (-15.37 = -14.76 + -0.61)


| Location | 5,641,985 – 5,642,086 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.24 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -14.97 |
| Energy contribution | -15.19 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
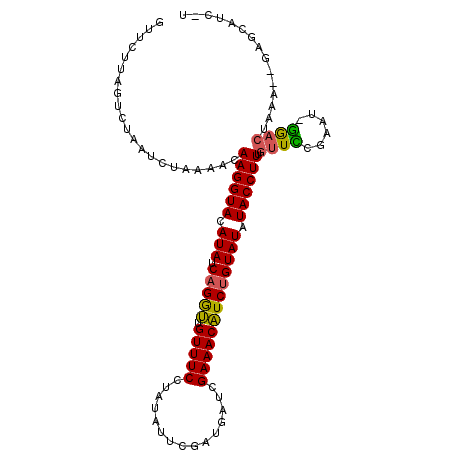
>2L_DroMel_CAF1 5641985 101 - 22407834 A-GACGCAC--UUUUGUCC-AAUCGGAACAAAGGUAUAUACAGAUGUUUCGAU---CGAAUACAGGAAACAACCUGAUAUUUACCUUGUUUUAGAUUAGACUAAGAAA .-......(--((..(((.-(((((((((((.((((.((((((.((((((...---.........))))))..))).))).))))))))))).)))).))).)))... ( -25.00) >DroVir_CAF1 125989 106 - 1 A-CAUGUGCUUAUCAGUUU-AAUCCAAUGAAAGGUAUAUACAGAGGUUUCAAUAAUCAAAUUUAAGAAACACUCUGAUAUGUACCUUGUUUCAGAUUAGAUUGUUGUU .-...........((((((-((((......((((((((((((((((((((..(((......))).))))).))))).))))))))))......))))))))))..... ( -32.40) >DroEre_CAF1 88237 104 - 1 A-GAUGCUC--UUUCGUCC-AUUCGGAACAAAGGUAUAUACAGAUGUUUCGAUCAUCCAAUACAGGAAACAACCUGAUAUGUACCUUGUUUUAGAUUAGACUAAGAAC .-.....((--((..(((.-(((..((((((.(((((((((((.((((((...............))))))..))).))))))))))))))..)))..))).)))).. ( -27.56) >DroWil_CAF1 126026 106 - 1 AGAAAACUC--UUUAGUCCUAUUCGAAACAAAGGUAGAUACAGAUGUUUCAGGCUUCAAAUAUACGAAACAACCUGAUAUGUACCUUGUUUUAGACUAGAAUUAGAAU .........--((((((.(((.(((((((((.((((.((((((.((((((...............))))))..))).))).))))))))))).)).))).)))))).. ( -25.06) >DroYak_CAF1 87666 104 - 1 A-GAUGCUC--UUUCGUCC-AUUCGGAACAAAGGUAUAUACAGAUGUUUCGAUCAUCCAAUAUAGGAAACAACCUGAUAUGUACCUUGUUUUAGAUUAGACCAAGAAC .-.....((--((..(((.-(((..((((((.(((((((((((.((((((...............))))))..))).))))))))))))))..)))..))).)))).. ( -27.36) >DroAna_CAF1 83618 104 - 1 A-GUAGCUC--UUUAGUUU-AAUCGAAACAAAGGUAGAUACAGAUGUUUCAAACUUUGAAUAUAGGAAACAACCAGAUAUGUACCUUGUUUUAGAUUAAAUCAAGAAC .-.....((--((..((((-(((((((((((.((((.(((((((.((.....))))))......(....).......))).))))))))))).)))))))).)))).. ( -22.70) >consensus A_GAUGCUC__UUUAGUCC_AAUCGAAACAAAGGUAUAUACAGAUGUUUCAAUCAUCAAAUAUAGGAAACAACCUGAUAUGUACCUUGUUUUAGAUUAGACUAAGAAC ...............(((....(((((((((.((((.((((((.((((((...............))))))..))).))).))))))))))).))...)))....... (-14.97 = -15.19 + 0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:25 2006