| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 611,163 – 611,505 |
| Length | 342 |
| Max. P | 0.996538 |

| Location | 611,163 – 611,277 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.07 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -16.19 |
| Energy contribution | -17.50 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 611163 114 + 22407834 ------AUGACGAUUUCCCUUUAGCCAUUCUCAACGCCUACUAAAUUGAGCCAGUUCCUAAUUGGCCAUCUUAUGACCGAAAUUAGUUCUAAUUUCGCACUACUCUGCAGGCAUAAACGA ------....((..................................((.((((((.....))))))))..(((((.(((((((((....)))))))(((......))).))))))).)). ( -21.80) >DroSec_CAF1 614 114 + 1 ------AUGUGGAUUUCCCUUUAGCCAUCCUCAACGCCUACAAAAUUAAGUCAGUUCUUAAUUGGCCACCUUACGAUCGAAAUUAGUUCUAAUUUCGCACUACUCUGCAGGCAUAAACGA ------.((.((((..(......)..)))).))..((((....(((((((......)))))))...............(((((((....)))))))(((......)))))))........ ( -21.00) >DroEre_CAF1 2201 120 + 1 GGUCGGCUAAGGAUUUGCCUUUAGCCAUCCUCAACGCCAAUAAAAUGAUGCCAGUUCUUAAUUGGCCAUCUUAUGAACGAAAUUAGUUCUAAUUUUGCAGCACUCUGCAGGCAUAAGCGA ((..((((((((......))))))))..)).....(((........(((((((((.....))))).))))....((((.......))))......(((((....))))))))........ ( -36.50) >DroYak_CAF1 2254 119 + 1 AGUCUGCUAUGGAUAUUCCU-UAGGCAUUCUCAGCGCCUAUUAAAUUAAGCCCGCUCUUAAUUGGCCAUCCUAUGAACGAAAUUAGUUCUAAUUUUGCACCACUCUGCAGGCAAAAAAGA .((((((...((((...((.-(((((.........)))))...(((((((......)))))))))..))))......((((((((....)))))))).........))))))........ ( -27.60) >consensus ______AUAAGGAUUUCCCUUUAGCCAUCCUCAACGCCUACAAAAUUAAGCCAGUUCUUAAUUGGCCAUCUUAUGAACGAAAUUAGUUCUAAUUUCGCACCACUCUGCAGGCAUAAACGA .......(((((((............)))))))..((((..........((((((.....))))))............(((((((....)))))))(((......)))))))........ (-16.19 = -17.50 + 1.31)

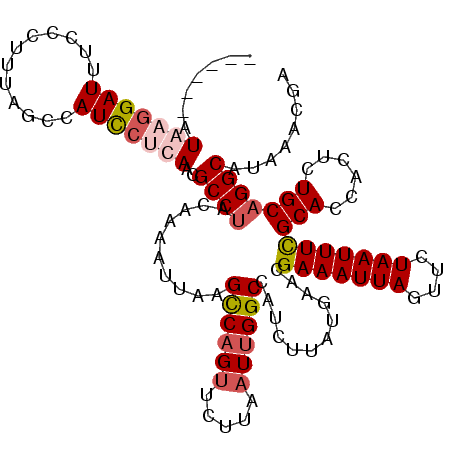
| Location | 611,197 – 611,311 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -29.41 |
| Energy contribution | -29.47 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.68 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 611197 114 + 22407834 CUAAAUUGAGCCAGUUCCUAAUUGGCCAUCUUAUGACCGAAAUUAGUUCUAAUUUCGCACUACUCUGCAGGCAUAAACGACUCUGCAGAAGCAGCU------ACAGUAGCUGCGAGGCAA .(((..((.((((((.....))))))))..))).....(((((((....)))))))((.((..((((((((..........)))))))).((((((------(...))))))).)))).. ( -36.00) >DroSec_CAF1 648 114 + 1 CAAAAUUAAGUCAGUUCUUAAUUGGCCACCUUACGAUCGAAAUUAGUUCUAAUUUCGCACUACUCUGCAGGCAUAAACGAGUCUGCAGAAGCAGCU------ACAGUAGCUGCGAAGCAA ...(((((((......)))))))...............(((((((....)))))))((.....(((((((((........))))))))).((((((------(...)))))))...)).. ( -36.20) >DroEre_CAF1 2241 120 + 1 UAAAAUGAUGCCAGUUCUUAAUUGGCCAUCUUAUGAACGAAAUUAGUUCUAAUUUUGCAGCACUCUGCAGGCAUAAGCGACUCUGCAGAAGCAGCUACUCGUACAGUAGCUGCGAAGCAA ......(((((((((.....))))).)))).......((((((((....))))))))..((..(((((((((....))....))))))).(((((((((.....)))))))))...)).. ( -40.80) >DroYak_CAF1 2293 114 + 1 UUAAAUUAAGCCCGCUCUUAAUUGGCCAUCCUAUGAACGAAAUUAGUUCUAAUUUUGCACCACUCUGCAGGCAAAAAAGACUCUGCAGAUGCAGCU------ACAGUAGCUGCGAGGCAA ...(((((((......))))))).(((..........((((((((....))))))))......((((((((..........))))))))(((((((------(...)))))))).))).. ( -32.20) >consensus CAAAAUUAAGCCAGUUCUUAAUUGGCCAUCUUAUGAACGAAAUUAGUUCUAAUUUCGCACCACUCUGCAGGCAUAAACGACUCUGCAGAAGCAGCU______ACAGUAGCUGCGAAGCAA .........((((((.....))))))............(((((((....)))))))((.....((((((((..........)))))))).((((((...........))))))...)).. (-29.41 = -29.47 + 0.06)

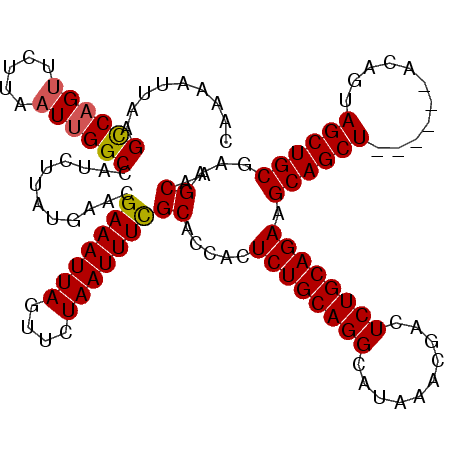
| Location | 611,197 – 611,311 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -25.84 |
| Energy contribution | -26.52 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 611197 114 - 22407834 UUGCCUCGCAGCUACUGU------AGCUGCUUCUGCAGAGUCGUUUAUGCCUGCAGAGUAGUGCGAAAUUAGAACUAAUUUCGGUCAUAAGAUGGCCAAUUAGGAACUGGCUCAAUUUAG ..(((..(((((((...)------))))))((((((((.((.......)))))))))).(((.((((((((....)))))))((((((...))))))......).))))))......... ( -37.00) >DroSec_CAF1 648 114 - 1 UUGCUUCGCAGCUACUGU------AGCUGCUUCUGCAGACUCGUUUAUGCCUGCAGAGUAGUGCGAAAUUAGAACUAAUUUCGAUCGUAAGGUGGCCAAUUAAGAACUGACUUAAUUUUG .(((...)))((((((..------.((((((.((((((.(........).)))))))))))).((((((((....)))))))).......)))))).(((((((......)))))))... ( -34.70) >DroEre_CAF1 2241 120 - 1 UUGCUUCGCAGCUACUGUACGAGUAGCUGCUUCUGCAGAGUCGCUUAUGCCUGCAGAGUGCUGCAAAAUUAGAACUAAUUUCGUUCAUAAGAUGGCCAAUUAAGAACUGGCAUCAUUUUA ((((..((((((((((.....)))))))))((((((((....((....)))))))))).)..)))).....((((.......)))).(((((((((((.........))))..))))))) ( -40.10) >DroYak_CAF1 2293 114 - 1 UUGCCUCGCAGCUACUGU------AGCUGCAUCUGCAGAGUCUUUUUUGCCUGCAGAGUGGUGCAAAAUUAGAACUAAUUUCGUUCAUAGGAUGGCCAAUUAAGAGCGGGCUUAAUUUAA ..((((((((((((...)------)))))).(((((((.((.......)))))))))((((.((.((((((....)))))).))))))..)).))).(((((((......)))))))... ( -33.30) >consensus UUGCCUCGCAGCUACUGU______AGCUGCUUCUGCAGAGUCGUUUAUGCCUGCAGAGUAGUGCAAAAUUAGAACUAAUUUCGUUCAUAAGAUGGCCAAUUAAGAACUGGCUUAAUUUAA ..(((((((((((...........)))))).(((((((............)))))))......((((((((....)))))))).......)).))).(((((((......)))))))... (-25.84 = -26.52 + 0.69)

| Location | 611,237 – 611,351 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -25.00 |
| Energy contribution | -25.50 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 611237 114 + 22407834 AAUUAGUUCUAAUUUCGCACUACUCUGCAGGCAUAAACGACUCUGCAGAAGCAGCU------ACAGUAGCUGCGAGGCAACUCAAACAUUUGUCAGCAACUUUGACUCCAAAUCGGCAUC ................((.....((((((((..........)))))))).((((((------(...)))))))(((....)))........(((((.....))))).........))... ( -30.50) >DroSec_CAF1 688 114 + 1 AAUUAGUUCUAAUUUCGCACUACUCUGCAGGCAUAAACGAGUCUGCAGAAGCAGCU------ACAGUAGCUGCGAAGCAACUCAAACAUUUGUCAGCAACUUUGACUCCAAAUCGGCAUC ................((.....(((((((((........))))))))).((((((------(...)))))))...)).................((...((((....))))...))... ( -30.80) >DroEre_CAF1 2281 120 + 1 AAUUAGUUCUAAUUUUGCAGCACUCUGCAGGCAUAAGCGACUCUGCAGAAGCAGCUACUCGUACAGUAGCUGCGAAGCAACUCAAACAUUUGUCAGCAACUUUGACUCCAAAUCGGCAUC ...............(((.((..(((((((((....))....))))))).(((((((((.....)))))))))...)).............(((((.....))))).........))).. ( -34.00) >DroYak_CAF1 2333 114 + 1 AAUUAGUUCUAAUUUUGCACCACUCUGCAGGCAAAAAAGACUCUGCAGAUGCAGCU------ACAGUAGCUGCGAGGCAACUCAAACAUUUGUCAGCAACUUUGACUCCAAAUCGGCAUC ...............(((.....((((((((..........)))))))).((((((------(...)))))))(((....)))........(((((.....))))).........))).. ( -30.50) >consensus AAUUAGUUCUAAUUUCGCACCACUCUGCAGGCAUAAACGACUCUGCAGAAGCAGCU______ACAGUAGCUGCGAAGCAACUCAAACAUUUGUCAGCAACUUUGACUCCAAAUCGGCAUC ................((.....((((((((..........)))))))).((((((...........))))))(((....)))........(((((.....))))).........))... (-25.00 = -25.50 + 0.50)

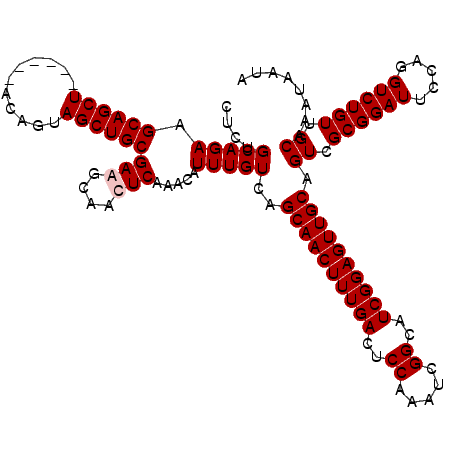
| Location | 611,277 – 611,391 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -40.45 |
| Consensus MFE | -35.53 |
| Energy contribution | -36.03 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 611277 114 + 22407834 CUCUGCAGAAGCAGCU------ACAGUAGCUGCGAGGCAACUCAAACAUUUGUCAGCAACUUUGACUCCAAAUCGGCAUCGGAGUUGCAGUCGCGGAUUCCAGGUCUGUUGCAAAUAAUA ....(((((.((((((------(...)))))))(((....))).....)))))..((((((((((..((.....))..)))))))))).((.((((((.....)))))).))........ ( -41.40) >DroSec_CAF1 728 114 + 1 GUCUGCAGAAGCAGCU------ACAGUAGCUGCGAAGCAACUCAAACAUUUGUCAGCAACUUUGACUCCAAAUCGGCAUCGGAGUUGCCGUCGCGGAUUCCAGGUCUGUUGCAAAUAAUA ((.(((....((((((------(...)))))))...)))))......(((((.(((((((((.((.(((....(((((.......)))))....))).)).)))).)))))))))).... ( -37.10) >DroEre_CAF1 2321 120 + 1 CUCUGCAGAAGCAGCUACUCGUACAGUAGCUGCGAAGCAACUCAAACAUUUGUCAGCAACUUUGACUCCAAAUCGGCAUCGGAGUUGCAGUCGCGGAUUCCAGGUCUGUUGCAAAUAAUA ...(((....(((((((((.....)))))))))...)))........((((((..((((((((((..((.....))..))))))))))....((((((.....)))))).)))))).... ( -41.10) >DroYak_CAF1 2373 114 + 1 CUCUGCAGAUGCAGCU------ACAGUAGCUGCGAGGCAACUCAAACAUUUGUCAGCAACUUUGACUCCAAAUCGGCAUCGGAGUUGCAGUCGCGGAUUCCAGGUCUGUUGCAAAUAAUA ....((((((((((((------(...)))))))(((....)))....))))))..((((((((((..((.....))..)))))))))).((.((((((.....)))))).))........ ( -42.20) >consensus CUCUGCAGAAGCAGCU______ACAGUAGCUGCGAAGCAACUCAAACAUUUGUCAGCAACUUUGACUCCAAAUCGGCAUCGGAGUUGCAGUCGCGGAUUCCAGGUCUGUUGCAAAUAAUA ....(((((.((((((...........))))))(((....))).....)))))..((((((((((..((.....))..)))))))))).((.((((((.....)))))).))........ (-35.53 = -36.03 + 0.50)

| Location | 611,277 – 611,391 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -38.62 |
| Consensus MFE | -35.25 |
| Energy contribution | -35.50 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 611277 114 - 22407834 UAUUAUUUGCAACAGACCUGGAAUCCGCGACUGCAACUCCGAUGCCGAUUUGGAGUCAAAGUUGCUGACAAAUGUUUGAGUUGCCUCGCAGCUACUGU------AGCUGCUUCUGCAGAG .....((((((.(((((......((.((((((...(((((((.......)))))))...)))))).)).....))))).........(((((((...)------))))))...)))))). ( -37.60) >DroSec_CAF1 728 114 - 1 UAUUAUUUGCAACAGACCUGGAAUCCGCGACGGCAACUCCGAUGCCGAUUUGGAGUCAAAGUUGCUGACAAAUGUUUGAGUUGCUUCGCAGCUACUGU------AGCUGCUUCUGCAGAC .....((((((.(((((......((.(((((....(((((((.......)))))))....))))).)).....))))).........(((((((...)------))))))...)))))). ( -35.60) >DroEre_CAF1 2321 120 - 1 UAUUAUUUGCAACAGACCUGGAAUCCGCGACUGCAACUCCGAUGCCGAUUUGGAGUCAAAGUUGCUGACAAAUGUUUGAGUUGCUUCGCAGCUACUGUACGAGUAGCUGCUUCUGCAGAG .....((((((.(((((......((.((((((...(((((((.......)))))))...)))))).)).....))))).........(((((((((.....)))))))))...)))))). ( -43.20) >DroYak_CAF1 2373 114 - 1 UAUUAUUUGCAACAGACCUGGAAUCCGCGACUGCAACUCCGAUGCCGAUUUGGAGUCAAAGUUGCUGACAAAUGUUUGAGUUGCCUCGCAGCUACUGU------AGCUGCAUCUGCAGAG .....((((((.(((((......((.((((((...(((((((.......)))))))...)))))).)).....))))).........(((((((...)------))))))...)))))). ( -38.10) >consensus UAUUAUUUGCAACAGACCUGGAAUCCGCGACUGCAACUCCGAUGCCGAUUUGGAGUCAAAGUUGCUGACAAAUGUUUGAGUUGCCUCGCAGCUACUGU______AGCUGCUUCUGCAGAG .....((((((.(((((......((.((((((...(((((((.......)))))))...)))))).)).....))))).........((((((...........))))))...)))))). (-35.25 = -35.50 + 0.25)

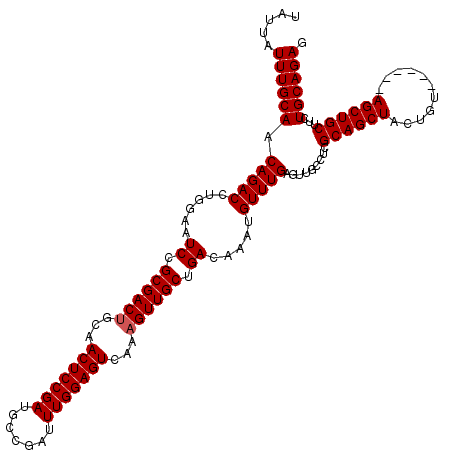
| Location | 611,311 – 611,431 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -36.30 |
| Energy contribution | -36.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 611311 120 + 22407834 CUCAAACAUUUGUCAGCAACUUUGACUCCAAAUCGGCAUCGGAGUUGCAGUCGCGGAUUCCAGGUCUGUUGCAAAUAAUAAAAAUCGGUAGCAAUUGCAAUUGUUGCUGGGCACAAAGGG ..........((((.((((((((((..((.....))..)))))))))).((.((((((.....)))))).)).............((((((((((....))))))))))))))....... ( -36.50) >DroSec_CAF1 762 120 + 1 CUCAAACAUUUGUCAGCAACUUUGACUCCAAAUCGGCAUCGGAGUUGCCGUCGCGGAUUCCAGGUCUGUUGCAAAUAAUAAAAAUCGGUAGCAAUUGCAAUUGUUGCUGGGCACAAAGGG ..........((((.(((((...((((...((((.((..(((.....)))..)).))))...)))).))))).............((((((((((....))))))))))))))....... ( -36.80) >DroEre_CAF1 2361 120 + 1 CUCAAACAUUUGUCAGCAACUUUGACUCCAAAUCGGCAUCGGAGUUGCAGUCGCGGAUUCCAGGUCUGUUGCAAAUAAUAAAAAUCGGUAGCAAUUGCAAUUGUUGCUGGGCACAAAGGG ..........((((.((((((((((..((.....))..)))))))))).((.((((((.....)))))).)).............((((((((((....))))))))))))))....... ( -36.50) >DroYak_CAF1 2407 120 + 1 CUCAAACAUUUGUCAGCAACUUUGACUCCAAAUCGGCAUCGGAGUUGCAGUCGCGGAUUCCAGGUCUGUUGCAAAUAAUAAAAAUCGGUAGCAAUUGCAAUUGUUGCUGGGCACAAAGGG ..........((((.((((((((((..((.....))..)))))))))).((.((((((.....)))))).)).............((((((((((....))))))))))))))....... ( -36.50) >consensus CUCAAACAUUUGUCAGCAACUUUGACUCCAAAUCGGCAUCGGAGUUGCAGUCGCGGAUUCCAGGUCUGUUGCAAAUAAUAAAAAUCGGUAGCAAUUGCAAUUGUUGCUGGGCACAAAGGG ..........((((.((((((((((..((.....))..)))))))))).((.((((((.....)))))).)).............((((((((((....))))))))))))))....... (-36.30 = -36.30 + 0.00)

| Location | 611,351 – 611,465 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.59 |
| Mean single sequence MFE | -39.58 |
| Consensus MFE | -38.97 |
| Energy contribution | -39.73 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 611351 114 + 22407834 GGAGUUGCAGUCGCGGAUUCCAGGUCUGUUGCAAAUAAUAAAAAUCGGUAGCAAUUGCAAUUGUUGCUGGGCACAAAGGGAAGCAAAGCGGUGGACUUGCGGAUCCA------AAUCCGA (((...((....))((((((((((((..((((............(((((((((((....)))))))))))((..........))...))))..)))))).)))))).------..))).. ( -41.70) >DroSec_CAF1 802 113 + 1 GGAGUUGCCGUCGCGGAUUCCAGGUCUGUUGCAAAUAAUAAAAAUCGGUAGCAAUUGCAAUUGUUGCUGGGCACAAAGGG-AGCAAAGCGGAGGACUUGCGGAUCCA------AGUCCGA (((.((((....))(((((((((((((.((((............(((((((((((....)))))))))))((.(.....)-.))...)))).))))))).)))))))------).))).. ( -40.00) >DroEre_CAF1 2401 120 + 1 GGAGUUGCAGUCGCGGAUUCCAGGUCUGUUGCAAAUAAUAAAAAUCGGUAGCAAUUGCAAUUGUUGCUGGGCACAAAGGGAAGCAAAGCGUUGGACUUGGGGAUCCAAAUCCAAAUCCGA (((...((....))(((((((((((((..(((............(((((((((((....)))))))))))((..........))...)))..)))))).)))))))...)))........ ( -38.30) >DroYak_CAF1 2447 114 + 1 GGAGUUGCAGUCGCGGAUUCCAGGUCUGUUGCAAAUAAUAAAAAUCGGUAGCAAUUGCAAUUGUUGCUGGGCACAAAGGGAAGCAAAGCGUUGGACUUGGGGAUCCA------AAUCCGA (((...((....))(((((((((((((..(((............(((((((((((....)))))))))))((..........))...)))..)))))).))))))).------..))).. ( -38.30) >consensus GGAGUUGCAGUCGCGGAUUCCAGGUCUGUUGCAAAUAAUAAAAAUCGGUAGCAAUUGCAAUUGUUGCUGGGCACAAAGGGAAGCAAAGCGGUGGACUUGCGGAUCCA______AAUCCGA (((...((....))((((((((((((((((((............(((((((((((....)))))))))))((..........))...)))))))))))).)))))).........))).. (-38.97 = -39.73 + 0.75)


| Location | 611,391 – 611,505 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.73 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -23.92 |
| Energy contribution | -24.17 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 611391 114 - 22407834 GAUCAACAUGUUGCACCACCGCACCACCAUGGCCACGGCCUCGGAUU------UGGAUCCGCAAGUCCACCGCUUUGCUUCCCUUUGUGCCCAGCAACAAUUGCAAUUGCUACCGAUUUU ((((....((((((......((((......(((....)))..(((((------((......)))))))..................))))...))))))...((....))....)))).. ( -27.80) >DroSec_CAF1 842 113 - 1 GAUAAACAUGUUGCACCACCGCACCGCCAUGGCCCCGGCCUCGGACU------UGGAUCCGCAAGUCCUCCGCUUUGCU-CCCUUUGUGCCCAGCAACAAUUGCAAUUGCUACCGAUUUU ........((((((......(((..((...(((....)))..(((((------((......)))))))...))..))).-.....((....))))))))...((....)).......... ( -28.20) >DroEre_CAF1 2441 120 - 1 GAUCAACAUGUUGCACCACCGCACCACCAUGGCCACGGCCUCGGAUUUGGAUUUGGAUCCCCAAGUCCAACGCUUUGCUUCCCUUUGUGCCCAGCAACAAUUGCAAUUGCUACCGAUUUU ((((....((((((......((((..((..(((....)))..))..((((((((((....))))))))))................))))...))))))...((....))....)))).. ( -35.10) >DroYak_CAF1 2487 114 - 1 GAUCAACAUGUUGCACCACCGCACCACCAUGGCCACGGCCUCGGAUU------UGGAUCCCCAAGUCCAACGCUUUGCUUCCCUUUGUGCCCAGCAACAAUUGCAAUUGCUACCGAUUUU ((((....((((((......((((......(((....)))..(((((------(((....))))))))..................))))...))))))...((....))....)))).. ( -31.00) >consensus GAUCAACAUGUUGCACCACCGCACCACCAUGGCCACGGCCUCGGAUU______UGGAUCCCCAAGUCCAACGCUUUGCUUCCCUUUGUGCCCAGCAACAAUUGCAAUUGCUACCGAUUUU ((((....((((((......((((..((..(((....)))..))..........((((......))))..................))))...))))))...((....))....)))).. (-23.92 = -24.17 + 0.25)

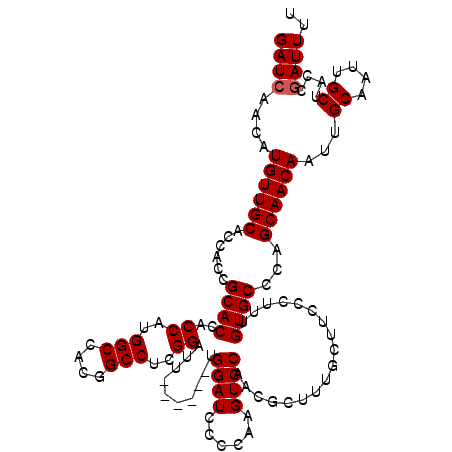
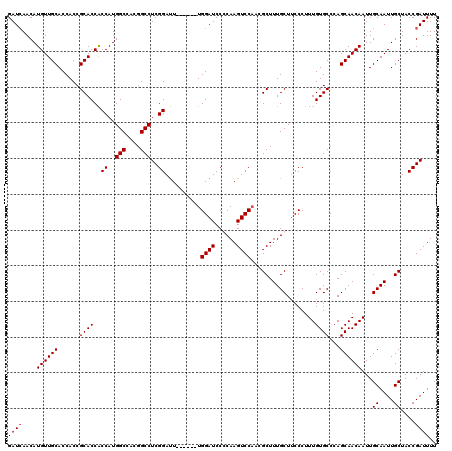
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:29:33 2006