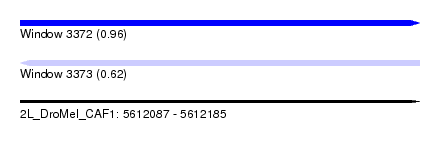
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,612,087 – 5,612,185 |
| Length | 98 |
| Max. P | 0.958246 |

| Location | 5,612,087 – 5,612,185 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.60 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -15.18 |
| Energy contribution | -15.32 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
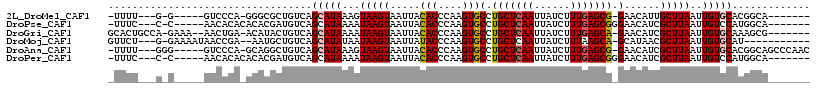
>2L_DroMel_CAF1 5612087 98 + 22407834 -UUUU---G-G-----GUCCCA-GGGCGCUGUCAGCAUAAAGUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCUUUGAGCG-GAACAUUGCUUAAUUGUGCACGGCA------- -....---.-.-----(((...-.)))(((((..((((((..((((((((..(((....))).((((((((......)))))))-)...)))))))).))))))))))).------- ( -33.70) >DroPse_CAF1 71677 100 + 1 -UUUC---C-C-----AACACACACACGAUGUCAGCAUAAAAUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCUUUGAGCGGGAACAUCGCUUAAUUGUCCAUGGCA------- -....---(-(-----((((......((((((....................(((....)))(((((((((......))))))))).))))))......)))...)))..------- ( -23.60) >DroGri_CAF1 65540 105 + 1 GCACUGCCA-GAAA--AACUGA-ACAUACUGUCAGCAUAAAAUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCUUUGAGCA-GAACAUCGCUUAAUUGUGCAAAGCG------- ((.....((-(...--..))).-...........((((((..(((((.....(((....))).((((((((......)))))))-)......))))).))))))...)).------- ( -25.80) >DroMoj_CAF1 100344 99 + 1 GUUCU---G-GAAAAUAACCGA--AAUGCUGUCAGCAUAUAAUAAGUAAUUAUACCCAAGUGCCUGCUCAAUUAUCUUUAAGCA-GCAUAACGCUUAAUUGUGCAU----------- ....(---(-(...........--.((((.....))))...((((....))))..))).((((.((((.((......)).))))-))))...((........))..----------- ( -16.60) >DroAna_CAF1 55978 106 + 1 -UUUU---GGG-----GUCCCA-GCAGGCUGUCAGCAUAAAGUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCUUUGAGCG-GAACAUCGCUUAAUUGUGCACGGCAGCCCAAC -..((---((.-----...)))-)..(((((((.((((((.((((....))))....(((((.((((((((......)))))))-).....)))))..))))))..))))))).... ( -38.80) >DroPer_CAF1 71720 100 + 1 -UUUC---C-C-----AACACACACACGAUGUCAGCAUAAAAUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCUUUGAGCGGGAACAUCGCUUAAUUGUCCAUGGCA------- -....---(-(-----((((......((((((....................(((....)))(((((((((......))))))))).))))))......)))...)))..------- ( -23.60) >consensus _UUUU___G_G_____AACCCA_ACACGCUGUCAGCAUAAAAUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCUUUGAGCG_GAACAUCGCUUAAUUGUGCAUGGCA_______ ..................................(((((...(((((.....(((....)))(.(((((((......))))))).)......)))))..)))))............. (-15.18 = -15.32 + 0.14)
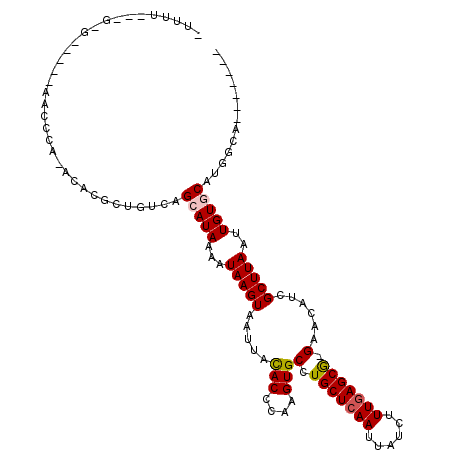
| Location | 5,612,087 – 5,612,185 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.60 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -16.85 |
| Energy contribution | -16.13 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5612087 98 - 22407834 -------UGCCGUGCACAAUUAAGCAAUGUUC-CGCUCAAAGAUAAUUGAGCAGGCACUUGGGUGUAAUUACUUACUUUAUGCUGACAGCGCCC-UGGGAC-----C-C---AAAA- -------.....(((........)))..((((-(((((((......)))))).(((.(((.(((((((.........))))))).).)).))).-.)))))-----.-.---....- ( -26.20) >DroPse_CAF1 71677 100 - 1 -------UGCCAUGGACAAUUAAGCGAUGUUCCCGCUCAAAGAUAAUUGAGCAGGCACUUGGGUGUAAUUACUUAUUUUAUGCUGACAUCGUGUGUGUGUU-----G-G---GAAA- -------..((...((((.....((((((((...((((((......)))))).((((..((((((....)))))).....)))))))))))).....))))-----.-)---)...- ( -29.20) >DroGri_CAF1 65540 105 - 1 -------CGCUUUGCACAAUUAAGCGAUGUUC-UGCUCAAAGAUAAUUGAGCAGGCACUUGGGUGUAAUUACUUAUUUUAUGCUGACAGUAUGU-UCAGUU--UUUC-UGGCAGUGC -------.((.((((.((..((((..((...(-(((((((......))))))))((((....)))).))..))))......(((((........-))))).--....-)))))).)) ( -27.10) >DroMoj_CAF1 100344 99 - 1 -----------AUGCACAAUUAAGCGUUAUGC-UGCUUAAAGAUAAUUGAGCAGGCACUUGGGUAUAAUUACUUAUUAUAUGCUGACAGCAUU--UCGGUUAUUUUC-C---AGAAC -----------.........(((.((..((((-(((((((......)))))).((((..((((((....)))))).....))))...))))).--.)).))).....-.---..... ( -21.80) >DroAna_CAF1 55978 106 - 1 GUUGGGCUGCCGUGCACAAUUAAGCGAUGUUC-CGCUCAAAGAUAAUUGAGCAGGCACUUGGGUGUAAUUACUUACUUUAUGCUGACAGCCUGC-UGGGAC-----CCC---AAAA- .(((((.(.((.....((((((((((......-))))......))))))(((((((...(.(((((((.........))))))).)..))))))-))).).-----)))---))..- ( -33.90) >DroPer_CAF1 71720 100 - 1 -------UGCCAUGGACAAUUAAGCGAUGUUCCCGCUCAAAGAUAAUUGAGCAGGCACUUGGGUGUAAUUACUUAUUUUAUGCUGACAUCGUGUGUGUGUU-----G-G---GAAA- -------..((...((((.....((((((((...((((((......)))))).((((..((((((....)))))).....)))))))))))).....))))-----.-)---)...- ( -29.20) >consensus _______UGCCAUGCACAAUUAAGCGAUGUUC_CGCUCAAAGAUAAUUGAGCAGGCACUUGGGUGUAAUUACUUAUUUUAUGCUGACAGCGUGU_UGGGUU_____C_C___AAAA_ ..........(((((...................((((((......)))))).((((..((((((....)))))).....))))....)))))........................ (-16.85 = -16.13 + -0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:15 2006