
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,610,461 – 5,610,553 |
| Length | 92 |
| Max. P | 0.582813 |

| Location | 5,610,461 – 5,610,553 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.03 |
| Mean single sequence MFE | -13.86 |
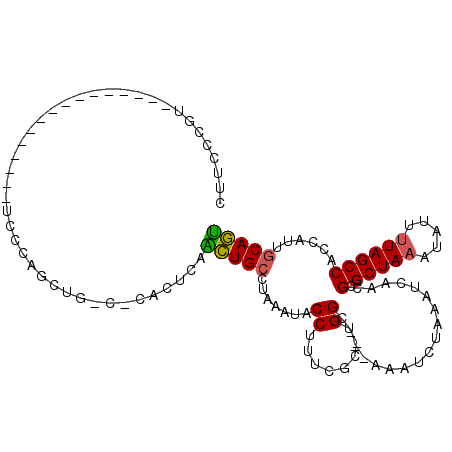
| Consensus MFE | -9.70 |
| Energy contribution | -9.37 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.23 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
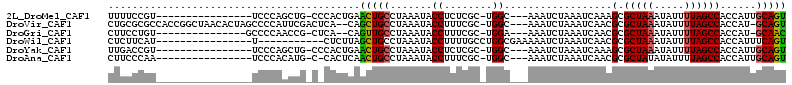
>2L_DroMel_CAF1 5610461 92 - 22407834 UUUUCCGU----------------UCCCAGCUG-CCCACUGAACUGCCUAAAUACCUCUCGC-UGGC---AAAUCUAAAUCAAAGCGCUAAAUAUUUUAGCCACCAUUGCAGU ........----------------.....((((-(.....((..((((..............-.)))---)..)).........(.(((((.....))))))......))))) ( -14.76) >DroVir_CAF1 80065 106 - 1 CUGCGCGCCACCGGCUAACACUAGCCCCAUUCGACUCA--CAGCUGCCUAAAUACCUUUCGC-UGGC---AAAUCUAAAUCAACGCGCUAAAUAUUUUAGCCACCAU-GCAGU (((((((.....(((((....)))))............--....((((..............-.)))---)............)))(((((.....)))))......-)))). ( -23.26) >DroGri_CAF1 63664 90 - 1 CUUCCUGU---------------GCCCCAACCG-CUCA--CAGUUGCCUAAAUACCUUUCGC-UGGA---AAAUCUAAAUCAACGCGCUAAAUAUUUUAGCCACCAU-GCAAC ....((((---------------(..(.....)-..))--)))((((.......((......-.)).---..............(.(((((.....)))))).....-)))). ( -11.40) >DroWil_CAF1 81459 86 - 1 CUCUUCAU----------------U-----------CUCUUAGCUGCCUAAAUACCUUUUGCCUGGCGAAAAAUCUAAAUCAACGCGCUAAAUAUUUUAGCCACCAUUUCAGU ........----------------.-----------......((((..........((((((...)))))).............(.(((((.....)))))).......)))) ( -8.90) >DroYak_CAF1 54828 92 - 1 UUGACCGU----------------UCCCAGCUG-CCCACUGAACUGCCUAAAUACCUCUCGC-UGGC---AAAUCUAAAUCAAAGCGCUAAAUAUUUUAGCCACCAUUGCAGU ........----------------.....((((-(.....((..((((..............-.)))---)..)).........(.(((((.....))))))......))))) ( -14.76) >DroAna_CAF1 54449 91 - 1 CUUCCCAA----------------UCCCACAUG-C-CACUCAACUGCCUAAAUACCUUUCGC-UGGC---AAAUCUAAAUCAACGCGCUAUAUAUUUUAGCCACCAUUGCAGU ........----------------.......((-(-........((((..............-.)))---).............(.((((.......)))))......))).. ( -10.06) >consensus CUUCCCGU________________UCCCAGCUG_C_CACUCAACUGCCUAAAUACCUUUCGC_UGGC___AAAUCUAAAUCAACGCGCUAAAUAUUUUAGCCACCAUUGCAGU ..........................................(((((.......((........))..................(.(((((.....))))))......))))) ( -9.70 = -9.37 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:13 2006