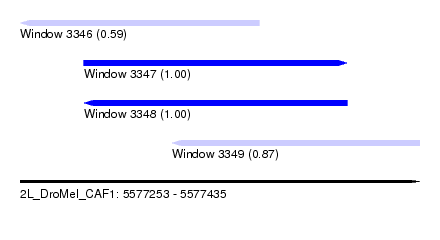
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,577,253 – 5,577,435 |
| Length | 182 |
| Max. P | 0.999955 |

| Location | 5,577,253 – 5,577,362 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.95 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -20.26 |
| Energy contribution | -21.26 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5577253 109 - 22407834 ACCUGUAUUAUUAGCCUGCAUGUAAUACUCGCAAAGUGCAUGCUUUUGACUUUGCCGCAUACGUUUCAAAGUCAUUAGGCGGCGC--ACUCGCAGAACAGAUG---------CUUUGACC ....(((((......((((.(((.......))).(((((.(((((.((((((((..((....))..))))))))..)))))..))--))).))))....))))---------)....... ( -29.80) >DroSec_CAF1 21288 111 - 1 ACCUGUAUUAUUAGCCUCCAUGUAAUACUCGCAAAGUGCAUGCUUUUGACUUUGCCGCAUACGUUUCAAAGUCAUUAGGCGGCGCACACUCGCAGAGCAGAUG---------CUCUGACC ....(((((((..........))))))).......((((.(((((.((((((((..((....))..))))))))..)))))..)))).....((((((....)---------)))))... ( -35.50) >DroSim_CAF1 22936 111 - 1 ACCUGUAUUAUUAGCCUCCAUGUAAUACUCGCAAAGUGCAUGCUUUUGACUUUGCCGCAUACGUUUCAAAGUCAUUAGGCGGCGCACACUUGCAGAGCAGAUG---------CUCUGACC ....(((((((..........)))))))..((((.((((.(((((.((((((((..((....))..))))))))..)))))..))))..))))(((((....)---------)))).... ( -36.10) >DroEre_CAF1 21233 118 - 1 ACGUGUAUUAUUAGCGUCCAUGUAAUACUCGCAAAGUGCAUGCUUUUCACUUUGCCGCAUACGUUUCAAAGUCAUUAGGCGGCGC--ACUCGCAGAGCAGAUGCUAGCCCAACUCUGACC ..........((((((((..((.....(((((..(((((.(((((...((((((..((....))..))))))....)))))..))--))).)).)))))))))))))............. ( -28.30) >DroYak_CAF1 21140 101 - 1 ACCUGUAUUAUUAGCUUCCAUGUAAUACUCGCAAAGUGCAUGCUUUUGACUUUGCCGCAUACGUUUCAAAGUCAUUAGGCG-------------------AUGCUAACCCAACUCUGACC ....(((((((..........))))))).........((((((((.((((((((..((....))..))))))))..)))).-------------------))))................ ( -22.40) >consensus ACCUGUAUUAUUAGCCUCCAUGUAAUACUCGCAAAGUGCAUGCUUUUGACUUUGCCGCAUACGUUUCAAAGUCAUUAGGCGGCGC__ACUCGCAGAGCAGAUG_________CUCUGACC ....(((((((..........))))))).......((((.(((((.((((((((..((....))..))))))))..)))))))))................................... (-20.26 = -21.26 + 1.00)

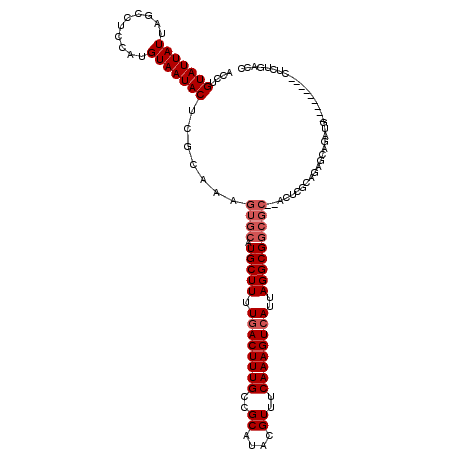
| Location | 5,577,282 – 5,577,402 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.42 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -30.44 |
| Energy contribution | -31.04 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5577282 120 + 22407834 GCCUAAUGACUUUGAAACGUAUGCGGCAAAGUCAAAAGCAUGCACUUUGCGAGUAUUACAUGCAGGCUAAUAAUACAGGUGAGCAUAAUAGCGACCUGCAACGCAAUAUGAAUGCUAAAU ......((((((((...((....)).))))))))..(((((.((..(((((.((((((...(....)...))))))((((..((......)).))))....)))))..)).))))).... ( -35.90) >DroSec_CAF1 21319 120 + 1 GCCUAAUGACUUUGAAACGUAUGCGGCAAAGUCAAAAGCAUGCACUUUGCGAGUAUUACAUGGAGGCUAAUAAUACAGGUGAGCAUAAUAGCGAGCUGCAACGCAAUAUGAAUGCUAAAU ......((((((((...((....)).))))))))..(((((.((..(((((.((((((..(((...))).)))))).(((..((......))..)))....)))))..)).))))).... ( -33.50) >DroSim_CAF1 22967 120 + 1 GCCUAAUGACUUUGAAACGUAUGCGGCAAAGUCAAAAGCAUGCACUUUGCGAGUAUUACAUGGAGGCUAAUAAUACAGGUGAGCAUAAUAGCGAGCUGCAACGCAAUAUGAAUGCUAAAU ......((((((((...((....)).))))))))..(((((.((..(((((.((((((..(((...))).)))))).(((..((......))..)))....)))))..)).))))).... ( -33.50) >DroEre_CAF1 21271 120 + 1 GCCUAAUGACUUUGAAACGUAUGCGGCAAAGUGAAAAGCAUGCACUUUGCGAGUAUUACAUGGACGCUAAUAAUACACGUGGGCAUAAUAGCGAGCUGCAACGCAAUAUGAAUGCUAAAU ......(.((((((...((....)).)))))).)..(((((.((..(((((.((((((..(((...))).))))))..((..((......))..)).....)))))..)).))))).... ( -27.60) >DroYak_CAF1 21161 120 + 1 GCCUAAUGACUUUGAAACGUAUGCGGCAAAGUCAAAAGCAUGCACUUUGCGAGUAUUACAUGGAAGCUAAUAAUACAGGUGAGCAUAAUAGCGAGCUGCAACGCAAUGUGAAUGCUAAAU ......((((((((...((....)).))))))))..(((((.(((.(((((.((((((...(....)...)))))).(((..((......))..)))....))))).))).))))).... ( -37.50) >consensus GCCUAAUGACUUUGAAACGUAUGCGGCAAAGUCAAAAGCAUGCACUUUGCGAGUAUUACAUGGAGGCUAAUAAUACAGGUGAGCAUAAUAGCGAGCUGCAACGCAAUAUGAAUGCUAAAU ......((((((((...((....)).))))))))..(((((.((..(((((.((((((...(....)...)))))).(((..((......))..)))....)))))..)).))))).... (-30.44 = -31.04 + 0.60)

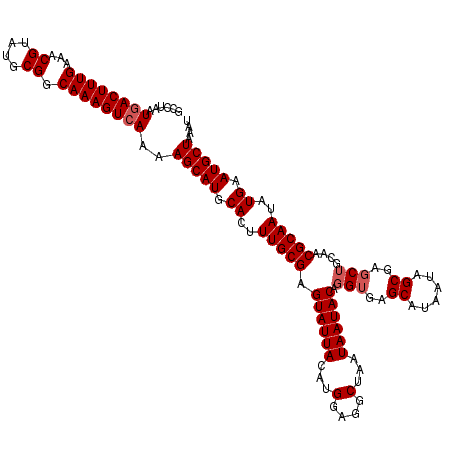
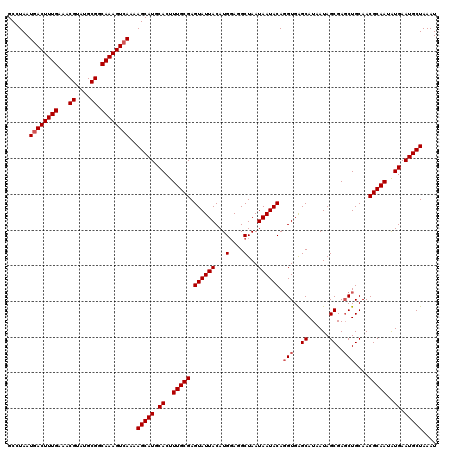
| Location | 5,577,282 – 5,577,402 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.42 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -32.20 |
| Energy contribution | -32.24 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.84 |
| SVM RNA-class probability | 0.999955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5577282 120 - 22407834 AUUUAGCAUUCAUAUUGCGUUGCAGGUCGCUAUUAUGCUCACCUGUAUUAUUAGCCUGCAUGUAAUACUCGCAAAGUGCAUGCUUUUGACUUUGCCGCAUACGUUUCAAAGUCAUUAGGC ....(((((.(((.(((((.(((((((....((.((((......)))).))..))))))).........))))).))).)))))..((((((((..((....))..))))))))...... ( -35.00) >DroSec_CAF1 21319 120 - 1 AUUUAGCAUUCAUAUUGCGUUGCAGCUCGCUAUUAUGCUCACCUGUAUUAUUAGCCUCCAUGUAAUACUCGCAAAGUGCAUGCUUUUGACUUUGCCGCAUACGUUUCAAAGUCAUUAGGC ....(((((.(((.((((((((((....((((..((((......))))...)))).....)))))....))))).))).)))))..((((((((..((....))..))))))))...... ( -33.10) >DroSim_CAF1 22967 120 - 1 AUUUAGCAUUCAUAUUGCGUUGCAGCUCGCUAUUAUGCUCACCUGUAUUAUUAGCCUCCAUGUAAUACUCGCAAAGUGCAUGCUUUUGACUUUGCCGCAUACGUUUCAAAGUCAUUAGGC ....(((((.(((.((((((((((....((((..((((......))))...)))).....)))))....))))).))).)))))..((((((((..((....))..))))))))...... ( -33.10) >DroEre_CAF1 21271 120 - 1 AUUUAGCAUUCAUAUUGCGUUGCAGCUCGCUAUUAUGCCCACGUGUAUUAUUAGCGUCCAUGUAAUACUCGCAAAGUGCAUGCUUUUCACUUUGCCGCAUACGUUUCAAAGUCAUUAGGC ....(((((.(((.((((((((((...(((((..((((......))))...)))))....)))))....))))).))).)))))....((((((..((....))..))))))........ ( -30.30) >DroYak_CAF1 21161 120 - 1 AUUUAGCAUUCACAUUGCGUUGCAGCUCGCUAUUAUGCUCACCUGUAUUAUUAGCUUCCAUGUAAUACUCGCAAAGUGCAUGCUUUUGACUUUGCCGCAUACGUUUCAAAGUCAUUAGGC ....(((((.(((.((((((((((....((((..((((......))))...)))).....)))))....))))).))).)))))..((((((((..((....))..))))))))...... ( -34.50) >consensus AUUUAGCAUUCAUAUUGCGUUGCAGCUCGCUAUUAUGCUCACCUGUAUUAUUAGCCUCCAUGUAAUACUCGCAAAGUGCAUGCUUUUGACUUUGCCGCAUACGUUUCAAAGUCAUUAGGC ....(((((.(((.((((((((((....((((..((((......))))...)))).....)))))....))))).))).)))))..((((((((..((....))..))))))))...... (-32.20 = -32.24 + 0.04)


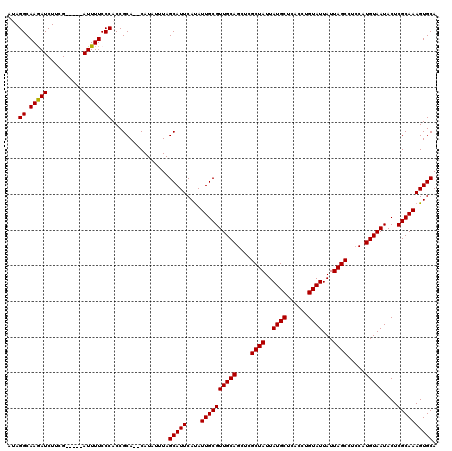
| Location | 5,577,322 – 5,577,435 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.84 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -21.14 |
| Energy contribution | -20.90 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5577322 113 - 22407834 AUAGGCAAGAUCUUCG-----AUUUUCCCACCGCA--CAUAUUUAGCAUUCAUAUUGCGUUGCAGGUCGCUAUUAUGCUCACCUGUAUUAUUAGCCUGCAUGUAAUACUCGCAAAGUGCA ...((.((((((...)-----))))).))...(((--(...(((.((.....((((((((.((((((....((.((((......)))).))..))))))))))))))...))))))))). ( -28.30) >DroSec_CAF1 21359 118 - 1 AUAGGCAAAAUCUUCGAUUCGAUUUUCCCACCGCA--CAUAUUUAGCAUUCAUAUUGCGUUGCAGCUCGCUAUUAUGCUCACCUGUAUUAUUAGCCUCCAUGUAAUACUCGCAAAGUGCA ...((.((((((........)))))).))...(((--(.......(((.......)))((((((....((((..((((......))))...)))).....)))))).........)))). ( -27.30) >DroSim_CAF1 23007 118 - 1 AUAGGCAAAAUCUUCGAUUCGAUUUUCCCACCGCA--CAUAUUUAGCAUUCAUAUUGCGUUGCAGCUCGCUAUUAUGCUCACCUGUAUUAUUAGCCUCCAUGUAAUACUCGCAAAGUGCA ...((.((((((........)))))).))...(((--(.......(((.......)))((((((....((((..((((......))))...)))).....)))))).........)))). ( -27.30) >DroEre_CAF1 21311 113 - 1 UGAGGCAAGAUCUUCG-----AUUUUCCCACCGCA--CAUAUUUAGCAUUCAUAUUGCGUUGCAGCUCGCUAUUAUGCCCACGUGUAUUAUUAGCGUCCAUGUAAUACUCGCAAAGUGCA ...((.((((((...)-----))))).))...(((--(.......(((.......)))((((((...(((((..((((......))))...)))))....)))))).........)))). ( -28.00) >DroYak_CAF1 21201 115 - 1 GGAGGCAAGAUCUUCG-----AUUUUCCCACCGCACACAUAUUUAGCAUUCACAUUGCGUUGCAGCUCGCUAUUAUGCUCACCUGUAUUAUUAGCUUCCAUGUAAUACUCGCAAAGUGCA ((.((.((((((...)-----))))).)).))((((.........(((.......)))((((((....((((..((((......))))...)))).....)))))).........)))). ( -28.60) >consensus AUAGGCAAGAUCUUCG_____AUUUUCCCACCGCA__CAUAUUUAGCAUUCAUAUUGCGUUGCAGCUCGCUAUUAUGCUCACCUGUAUUAUUAGCCUCCAUGUAAUACUCGCAAAGUGCA ...((.(((((..........))))).))................(((((....((((((((((....((((..((((......))))...)))).....)))))....)))))))))). (-21.14 = -20.90 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:51 2006