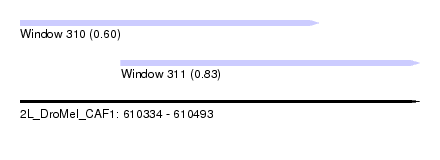
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 610,334 – 610,493 |
| Length | 159 |
| Max. P | 0.826593 |

| Location | 610,334 – 610,453 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.43 |
| Mean single sequence MFE | -48.21 |
| Consensus MFE | -42.70 |
| Energy contribution | -42.71 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 610334 119 + 22407834 CGGCAACAUUGACGUGUGUGGCAUGUGGGCAAAUUUGUUGUUGCCGC-UGGAGAUGUUGCUAGUCGCUGCGGUUGCAAUGCUGCCGUUGUCAUUGGCCGAGGAAACAAUGCGGCGGCGCU .(((((((((..((...((((((....((((....))))..))))))-))..))))))))).((((((((((((((((((....))))))....))))..(....)...))))))))... ( -47.60) >DroEre_CAF1 1371 119 + 1 CGGCAACAUUGACGUGUGUGGCAUGUGGGCAAAUUUGUUGUUGCUGC-UGGAGAUGCUGCUAGUUGCUGCGGCUGCAAUGUGGCCGUUGUCAUUGGCCGCGGAAACAAUGCGGCGGCGCU .((((((....(((((.....))))).((((.((((.(.((....))-.).))))..)))).))))))((.((((((..(((((((.......)))))))(....)..)))))).))... ( -47.80) >DroYak_CAF1 1380 120 + 1 CGGCAACAUUGACGUGUGUGGCAUGUGGGCAAAUUUGUUGUUGCUGCCUGAAGAUGCUGCUAGUUGCUGCGGUUGCAAUGUGGCCGUUGUCAUUGGCCGCGGCAACAAUGCGGCGGCGCU .((((.((((.(((((.....)))))(((((.............)))))...)))).)))).((((((((.(((((...(((((((.......))))))).)))))...))))))))... ( -49.22) >consensus CGGCAACAUUGACGUGUGUGGCAUGUGGGCAAAUUUGUUGUUGCUGC_UGGAGAUGCUGCUAGUUGCUGCGGUUGCAAUGUGGCCGUUGUCAUUGGCCGCGGAAACAAUGCGGCGGCGCU .((((.((((..((...((((((....((((....))))..)))))).))..)))).)))).(((((((((.......((((((((.......)))))))).......)))))))))... (-42.70 = -42.71 + 0.01)

| Location | 610,374 – 610,493 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.31 |
| Mean single sequence MFE | -46.50 |
| Consensus MFE | -40.33 |
| Energy contribution | -40.33 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 610374 119 + 22407834 UUGCCGC-UGGAGAUGUUGCUAGUCGCUGCGGUUGCAAUGCUGCCGUUGUCAUUGGCCGAGGAAACAAUGCGGCGGCGCUGCAAUUUAAAUAAGUGCCCGGCAACAGAGAACAUUGAACA ....((.-((....((((((..(.((((..(((((((.((((((((....(((((.(....)...))))))))))))).)))))))......)))).)..)))))).....)).)).... ( -40.50) >DroEre_CAF1 1411 119 + 1 UUGCUGC-UGGAGAUGCUGCUAGUUGCUGCGGCUGCAAUGUGGCCGUUGUCAUUGGCCGCGGAAACAAUGCGGCGGCGCUGCAAUUUAAAUAAGUGCCCGGCAACAGGGAACAUCGAACA .......-....((((.(.((.(((((((..((((((..(((((((.......)))))))(....)..))))))((((((............))))))))))))).)).).))))..... ( -49.20) >DroYak_CAF1 1420 120 + 1 UUGCUGCCUGAAGAUGCUGCUAGUUGCUGCGGUUGCAAUGUGGCCGUUGUCAUUGGCCGCGGCAACAAUGCGGCGGCGCUGCAAUUUAAAUAAGUGCCCGGCAACAGAGAACAUUGAACA (((.((((.......(((((..((((((((((((((((((....))))))....))))))))))))...)))))((((((............)))))).)))).)))............. ( -49.80) >consensus UUGCUGC_UGGAGAUGCUGCUAGUUGCUGCGGUUGCAAUGUGGCCGUUGUCAUUGGCCGCGGAAACAAUGCGGCGGCGCUGCAAUUUAAAUAAGUGCCCGGCAACAGAGAACAUUGAACA ((((((.........(((((..(((.((((((((((((((....))))))....)))))))).)))...)))))((((((............))))))))))))................ (-40.33 = -40.33 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:29:21 2006