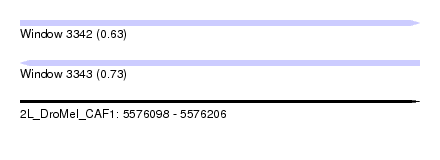
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,576,098 – 5,576,206 |
| Length | 108 |
| Max. P | 0.730194 |

| Location | 5,576,098 – 5,576,206 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.26 |
| Mean single sequence MFE | -41.64 |
| Consensus MFE | -22.14 |
| Energy contribution | -22.26 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5576098 108 + 22407834 -------CUGGAGGAUUUCCUGCAGUUCCCAGCCACUUCAGCGUCGACCACAUAAAGUGUCAUUAGCUCGAGUGGCGAAGCGACUGGCUUGGUGGACCAGGAGCAGGAUUCGGCU----- -------(((.(((....))).))).(((..(((((((.(((...(((.((.....)))))....))).)))))))...((..((((((....)).))))..)).))).......----- ( -41.00) >DroSec_CAF1 20125 111 + 1 -------CUGGAGGACUUCCCGCAAUUCCCAGCCACUUCAGCCUCGACCACAUAAAGUGUCAUUAGCUCGAGUGGCGAGGCGACUGCCUUGGUGGACCAGGAGCAGGAUUUGGCUGGC-- -------.((..((....))..))....(((((((..((.((((((.((((.(..(((.......)))..)))))))))))..(((((((((....))))).))))))..))))))).-- ( -49.00) >DroSim_CAF1 21793 112 + 1 -------CUGGAGGACUUCCCGCAGUUCCCAGCCACUUCUGCCUCGACCACAUAAAGUGUCAUUAGCUCGAGUGGCGAGGCGACUGCCUUGGUGGAC-AGGAGCAGGAUUCGGCUGGAGC -------(((..((....))..)))...((((((.....(((((((.((((.(..(((.......)))..)))))))))))).((((((((.....)-))).)))).....))))))... ( -46.30) >DroEre_CAF1 20046 90 + 1 -----------------UCCCGCAGUUCCCAGCCACUUCAGCCUCGACCACAUAAAGUGUCAUUAGCUCGAGUGGCGAGCAGACCA--------GGCCAGGAGCGGGAUUCGGCU----- -----------------((((((.((((...(((((((.(((...(((.((.....)))))....))).)))))))))))...((.--------.....)).)))))).......----- ( -33.90) >DroYak_CAF1 19948 102 + 1 AGCAGUGCUGAAGGACCUCCCGCAGUUCCCAGCCACUUCAGCCUCGGCCACAUAAAGUGUCAUUAGCUCGAGUGGCGAGGAGACC-------------AGGAGCAGCAUUCGGCU----- ......((((((((.....))((.(((((..(((((((.(((....(.(((.....))).)....))).)))))))(.(....))-------------.))))).)).)))))).----- ( -38.00) >consensus _______CUGGAGGACUUCCCGCAGUUCCCAGCCACUUCAGCCUCGACCACAUAAAGUGUCAUUAGCUCGAGUGGCGAGGCGACUG_CUUGGUGGACCAGGAGCAGGAUUCGGCU_____ ...................(((..(((((..(((((((.(((...(((.((.....)))))....))).)))))))..(....)...............)))))......)))....... (-22.14 = -22.26 + 0.12)



| Location | 5,576,098 – 5,576,206 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.26 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -27.74 |
| Energy contribution | -29.30 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5576098 108 - 22407834 -----AGCCGAAUCCUGCUCCUGGUCCACCAAGCCAGUCGCUUCGCCACUCGAGCUAAUGACACUUUAUGUGGUCGACGCUGAAGUGGCUGGGAACUGCAGGAAAUCCUCCAG------- -----.......((((((...(((....)))..(((((((((((((...((((.(..((((....))))..).)))).)).))))))))))).....))))))..........------- ( -37.80) >DroSec_CAF1 20125 111 - 1 --GCCAGCCAAAUCCUGCUCCUGGUCCACCAAGGCAGUCGCCUCGCCACUCGAGCUAAUGACACUUUAUGUGGUCGAGGCUGAAGUGGCUGGGAAUUGCGGGAAGUCCUCCAG------- --.(((((((..(((((((..(((....))).)))))..((((((((((..(((.........)))...)))).)))))).))..)))))))....((.(((....))).)).------- ( -45.50) >DroSim_CAF1 21793 112 - 1 GCUCCAGCCGAAUCCUGCUCCU-GUCCACCAAGGCAGUCGCCUCGCCACUCGAGCUAAUGACACUUUAUGUGGUCGAGGCAGAAGUGGCUGGGAACUGCGGGAAGUCCUCCAG------- ..((((((((..(((((((..(-(.....)).)))))..((((((((((..(((.........)))...)))).)))))).))..))))))))..(((.(((....))).)))------- ( -43.80) >DroEre_CAF1 20046 90 - 1 -----AGCCGAAUCCCGCUCCUGGCC--------UGGUCUGCUCGCCACUCGAGCUAAUGACACUUUAUGUGGUCGAGGCUGAAGUGGCUGGGAACUGCGGGA----------------- -----.......(((((((((..(((--------...((.(((((((((..(((.........)))...)))).))).)).))...)))..)))...))))))----------------- ( -38.10) >DroYak_CAF1 19948 102 - 1 -----AGCCGAAUGCUGCUCCU-------------GGUCUCCUCGCCACUCGAGCUAAUGACACUUUAUGUGGCCGAGGCUGAAGUGGCUGGGAACUGCGGGAGGUCCUUCAGCACUGCU -----(((.(..((((((((((-------------(...((((.((((((..((((.....(((.....))).....))))..)))))).))))....))))))......)))))).))) ( -38.80) >consensus _____AGCCGAAUCCUGCUCCUGGUCCACCAAG_CAGUCGCCUCGCCACUCGAGCUAAUGACACUUUAUGUGGUCGAGGCUGAAGUGGCUGGGAACUGCGGGAAGUCCUCCAG_______ ............((((((((((((((...........((((((((((((..(((.........)))...)))).)))))).))...))))))))...))))))................. (-27.74 = -29.30 + 1.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:45 2006