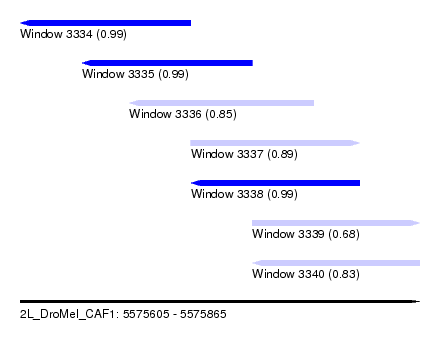
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,575,605 – 5,575,865 |
| Length | 260 |
| Max. P | 0.992601 |

| Location | 5,575,605 – 5,575,716 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.54 |
| Mean single sequence MFE | -21.02 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.52 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5575605 111 - 22407834 GCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCACACAAUGCAAGUGGAAAAUAAAU---------AAAAAAAAUCUUUACUCAACCCAACCCAACCACUGUUGUCCGCACAUGA .(((((.(((((...(((...........((((((.(((.....)))..))))))......(---------(((.......))))...................))).))))).))))). ( -21.70) >DroSec_CAF1 19653 106 - 1 GCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCA--CAAUGCAAGUGGAAAAUAAAU---------AAAAAAAAUCUUUACUCAACCCAACCCAA---CUGUUUUCCGCACAUGA .(((((.((((....(((...........((((((.((.--....))..))))))......(---------(((.......))))...............---.)))..)))).))))). ( -17.10) >DroSim_CAF1 21305 106 - 1 GCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCA--CAAUGCAAGUGGAAAAUAAAU---------AAAAAAAAUCUUUACUCAACCCAACCCAA---CUGUUGUCCGCACAUGA .(((((.(((((...(((...........((((((.((.--....))..))))))......(---------(((.......))))...............---.))).))))).))))). ( -21.30) >DroEre_CAF1 19573 99 - 1 GCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCA--CAAUGCACGUGGAAAAUAAAU---------ACAAAAAAUCUUUACUCAACC-------A---CUGUUGUCCGCACAUAA ..((((.(((((.................((((((.((.--....))..)))))).......---------.................(((.-------.---..)))))))).)))).. ( -18.50) >DroYak_CAF1 19414 108 - 1 GCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCA--CAAUGCAAGUGGAAAAUAAAUAAAGUGGAAACAAAAAAUCUUUACUCAACC-------A---CUGUUGUCCGCACAUGA .(((((.(((((...((............((((((.((.--....))..)))))).........(((((.....................))-------)---)))).))))).))))). ( -26.50) >consensus GCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCA__CAAUGCAAGUGGAAAAUAAAU_________AAAAAAAAUCUUUACUCAACCCAACCCAA___CUGUUGUCCGCACAUGA .(((((.(((((...(((...........((((((.(((.....)))..))))))...................(((....)))....................))).))))).))))). (-19.12 = -19.52 + 0.40)


| Location | 5,575,645 – 5,575,756 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.82 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -25.18 |
| Energy contribution | -25.58 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5575645 111 - 22407834 AUCAUUUGCAUCUGGCUAUAUCUCCAUUGAAGUGGAUGAGGCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCACACAAUGCAAGUGGAAAAUAAAU---------AAAAAAAAU .((((((((((.((((....(((((((....))))).))((((.(...((..((((......))))..)).))))))).))..)))))))))).........---------......... ( -26.50) >DroSec_CAF1 19690 109 - 1 AUCAUUUGCAUCUGGCUAUAUCUCCAUUGAAGUGGGUGAGGCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCA--CAAUGCAAGUGGAAAAUAAAU---------AAAAAAAAU .((((((((((.((((.......((((....))))(((((((.(....).)))))))................))))..--..)))))))))).........---------......... ( -28.20) >DroSim_CAF1 21342 109 - 1 AUCAUUUGCAUCUGGCUAUAUCUCCAUUGAAGUGGGUGAGGCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCA--CAAUGCAAGUGGAAAAUAAAU---------AAAAAAAAU .((((((((((.((((.......((((....))))(((((((.(....).)))))))................))))..--..)))))))))).........---------......... ( -28.20) >DroEre_CAF1 19603 107 - 1 AUCAUUUGCAUCUGGCUA--UCUCCAUUGAAGUGGGUGAGGCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCA--CAAUGCACGUGGAAAAUAAAU---------ACAAAAAAU .((((.(((((.((((..--...((((....))))(((((((.(....).)))))))................))))..--..))))).)))).........---------......... ( -25.00) >DroYak_CAF1 19444 116 - 1 AUCAUUUGCAUCUGGCUA--UCUCCAUUGAAGUGGGUGAGGCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCA--CAAUGCAAGUGGAAAAUAAAUAAAGUGGAAACAAAAAAU .((((((((((.((((..--...((((....))))(((((((.(....).)))))))................))))..--..)))))))))).............((....))...... ( -30.60) >consensus AUCAUUUGCAUCUGGCUAUAUCUCCAUUGAAGUGGGUGAGGCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCA__CAAUGCAAGUGGAAAAUAAAU_________AAAAAAAAU .((((((((((.((((.......((((....))))(((((((.(....).)))))))................))))......))))))))))........................... (-25.18 = -25.58 + 0.40)


| Location | 5,575,676 – 5,575,796 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.95 |
| Mean single sequence MFE | -33.49 |
| Consensus MFE | -31.50 |
| Energy contribution | -31.62 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5575676 120 - 22407834 AUAGCUAUAUUAAAGUUGUAGCUUGCAGGCCAUUAGGCCUAUCAUUUGCAUCUGGCUAUAUCUCCAUUGAAGUGGAUGAGGCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCAC ..(((((((.......)))))))((((((((....))))).(((((..(.(((((........)))..)).)..)))))((((.(...((..((((......))))..)).)))))))). ( -34.20) >DroSec_CAF1 19720 119 - 1 AUAGCUAUAUUAAAGUUGUAGCUUGCAGGCCAUUAGGCCUAUCAUUUGCAUCUGGCUAUAUCUCCAUUGAAGUGGGUGAGGCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCA- ..(((((((.......)))))))((((((((....))))).............(((.......((((....))))(((((((.(....).)))))))................))))))- ( -34.80) >DroSim_CAF1 21372 119 - 1 AUAGCUAUAUUAAAGUUGUAGCUUGCAGGCCAUUAGGCCUAUCAUUUGCAUCUGGCUAUAUCUCCAUUGAAGUGGGUGAGGCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCA- ..(((((((.......)))))))((((((((....))))).............(((.......((((....))))(((((((.(....).)))))))................))))))- ( -34.80) >DroEre_CAF1 19633 114 - 1 ---GCUAUAUUAAAGUUGUGCCGUGCAGGCCAUUAGGCCUAUCAUUUGCAUCUGGCUA--UCUCCAUUGAAGUGGGUGAGGCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCA- ---......((((((((((((((..((.(((....((((..............)))).--(((((((....))))).))))).))..)))....)))))))))))..............- ( -32.44) >DroYak_CAF1 19483 114 - 1 ---GCUGCAUUAAAGUUGUACCUUGCAGGCCAUUAGGCCUAUCAUUUGCAUCUGGCUA--UCUCCAUUGAAGUGGGUGAGGCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCA- ---...((.....((.((((...((.(((((....)))))..))..)))).))(((..--...((((....))))(((((((.(....).)))))))................))))).- ( -31.20) >consensus AUAGCUAUAUUAAAGUUGUAGCUUGCAGGCCAUUAGGCCUAUCAUUUGCAUCUGGCUAUAUCUCCAUUGAAGUGGGUGAGGCGUGUCCGGGCUUUACAAUUUUAAACACUUCCGCCGCA_ ...((((((.......)))))).((((((((....))))).............(((.......((((....))))(((((((.(....).)))))))................)))))). (-31.50 = -31.62 + 0.12)

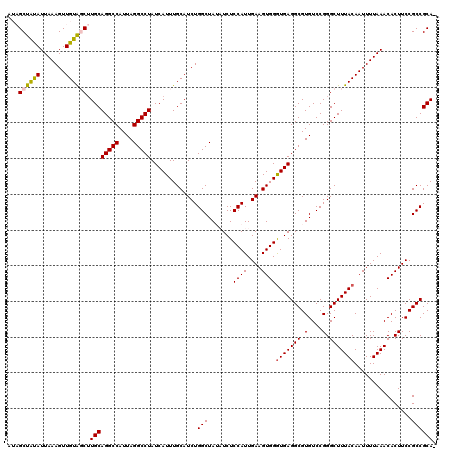
| Location | 5,575,716 – 5,575,826 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.32 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -20.40 |
| Energy contribution | -20.44 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5575716 110 + 22407834 CUCAUCCACUUCAAUGGAGAUAUAGCCAGAUGCAAAUGAUAGGCCUAAUGGCCUGCAAGCUACAACUUUAAUAUAGCUAUUGCUAUUGCUAUUGCUGGCAAAUG----------GCUUUA .((.((((......))))))...(((((...((((.((.((((((....))))))))(((((...........))))).))))..((((((....)))))).))----------)))... ( -32.30) >DroSec_CAF1 19759 110 + 1 CUCACCCACUUCAAUGGAGAUAUAGCCAGAUGCAAAUGAUAGGCCUAAUGGCCUGCAAGCUACAACUUUAAUAUAGCUAUUGCUAUUGCCAUUGCUGGCAAAUG----------GCUUUA .....(((......)))......(((((...((((.((.((((((....))))))))(((((...........))))).))))..((((((....)))))).))----------)))... ( -32.70) >DroSim_CAF1 21411 110 + 1 CUCACCCACUUCAAUGGAGAUAUAGCCAGAUGCAAAUGAUAGGCCUAAUGGCCUGCAAGCUACAACUUUAAUAUAGCUAUUGCUAUUGCCAUUGCUGGCAAAUG----------GCUUUA .....(((......)))......(((((...((((.((.((((((....))))))))(((((...........))))).))))..((((((....)))))).))----------)))... ( -32.70) >DroEre_CAF1 19672 96 + 1 CUCACCCACUUCAAUGGAGA--UAGCCAGAUGCAAAUGAUAGGCCUAAUGGCCUGCACGGCACAACUUUAAUAUAGC------------CAUUGCUGGCAAAUG----------GCUUUA .....(((......)))...--.(((((..(((...((.((((((....))))))))(((((...((.......)).------------...))))))))..))----------)))... ( -28.30) >DroYak_CAF1 19522 106 + 1 CUCACCCACUUCAAUGGAGA--UAGCCAGAUGCAAAUGAUAGGCCUAAUGGCCUGCAAGGUACAACUUUAAUGCAGC------------CAUUGCUGGCAAAUGGGCAAAGUGGGCUUUA ....((((((((......))--..(((...((((...(.((((((....)))))))((((.....))))..))))((------------((....)))).....)))..))))))..... ( -33.60) >consensus CUCACCCACUUCAAUGGAGAUAUAGCCAGAUGCAAAUGAUAGGCCUAAUGGCCUGCAAGCUACAACUUUAAUAUAGCUAUUGCUAUUGCCAUUGCUGGCAAAUG__________GCUUUA .....(((......))).......(((((.....((((.((((((....)))))).(((......))).....................)))).)))))..................... (-20.40 = -20.44 + 0.04)

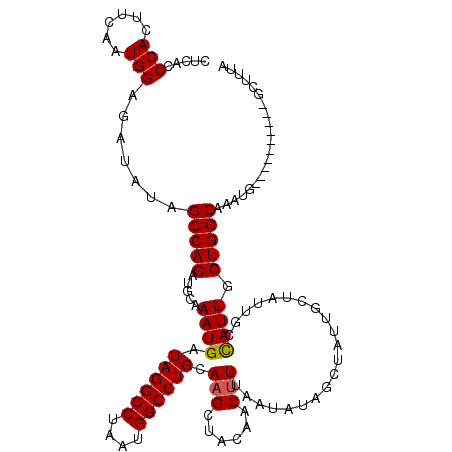

| Location | 5,575,716 – 5,575,826 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.32 |
| Mean single sequence MFE | -37.13 |
| Consensus MFE | -25.74 |
| Energy contribution | -25.50 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.82 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5575716 110 - 22407834 UAAAGC----------CAUUUGCCAGCAAUAGCAAUAGCAAUAGCUAUAUUAAAGUUGUAGCUUGCAGGCCAUUAGGCCUAUCAUUUGCAUCUGGCUAUAUCUCCAUUGAAGUGGAUGAG ...(((----------((..(((..((....))....(((..(((((((.......))))))))))(((((....))))).......)))..)))))...(((((((....))))).)). ( -36.60) >DroSec_CAF1 19759 110 - 1 UAAAGC----------CAUUUGCCAGCAAUGGCAAUAGCAAUAGCUAUAUUAAAGUUGUAGCUUGCAGGCCAUUAGGCCUAUCAUUUGCAUCUGGCUAUAUCUCCAUUGAAGUGGGUGAG ...(((----------((.((((((....))))))..((((.(((((((.......)))))))...(((((....))))).....))))...)))))...(((((((....))))).)). ( -39.50) >DroSim_CAF1 21411 110 - 1 UAAAGC----------CAUUUGCCAGCAAUGGCAAUAGCAAUAGCUAUAUUAAAGUUGUAGCUUGCAGGCCAUUAGGCCUAUCAUUUGCAUCUGGCUAUAUCUCCAUUGAAGUGGGUGAG ...(((----------((.((((((....))))))..((((.(((((((.......)))))))...(((((....))))).....))))...)))))...(((((((....))))).)). ( -39.50) >DroEre_CAF1 19672 96 - 1 UAAAGC----------CAUUUGCCAGCAAUG------------GCUAUAUUAAAGUUGUGCCGUGCAGGCCAUUAGGCCUAUCAUUUGCAUCUGGCUA--UCUCCAUUGAAGUGGGUGAG ...(((----------((..(((..(((.((------------((.(((.......))))))))))(((((....))))).......)))..))))).--(((((((....))))).)). ( -33.80) >DroYak_CAF1 19522 106 - 1 UAAAGCCCACUUUGCCCAUUUGCCAGCAAUG------------GCUGCAUUAAAGUUGUACCUUGCAGGCCAUUAGGCCUAUCAUUUGCAUCUGGCUA--UCUCCAUUGAAGUGGGUGAG ....(((((((((........((((((((.(------------(.((((.......))))))))))(((((....)))))............))))..--........)))))))))... ( -36.27) >consensus UAAAGC__________CAUUUGCCAGCAAUGGCAAUAGCAAUAGCUAUAUUAAAGUUGUAGCUUGCAGGCCAUUAGGCCUAUCAUUUGCAUCUGGCUAUAUCUCCAUUGAAGUGGGUGAG .....................((((((((..............((((((.......))))))....(((((....))))).....))))...))))....(((((((....))))).)). (-25.74 = -25.50 + -0.24)

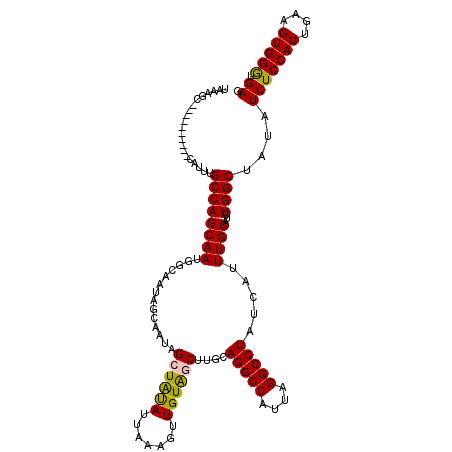

| Location | 5,575,756 – 5,575,865 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -14.84 |
| Energy contribution | -15.40 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5575756 109 + 22407834 AGGCCUAAUGGCCUGCAAGCUACAACUUUAAUAUAGCUAUUGCUAUUGCUAUUGCUGGCAAAUG----------GCUUUAAGCACCAGCACUCGAUAGAAACAUGG-CCGAGUUCAGGCA (((((....)))))....(((..(((((....(((((....))))).(((((((((((......----------((.....)).)))))).((....))...))))-).)))))..))). ( -30.80) >DroSec_CAF1 19799 109 + 1 AGGCCUAAUGGCCUGCAAGCUACAACUUUAAUAUAGCUAUUGCUAUUGCCAUUGCUGGCAAAUG----------GCUUUAAACACCAGCACUCGAUGGAAACAUGG-CCGAGUUCAGACA (((((....)))))((.(((((...........)))))...((((((((((....)))).))))----------))...........))((((((((....)))..-.)))))....... ( -33.80) >DroSim_CAF1 21451 109 + 1 AGGCCUAAUGGCCUGCAAGCUACAACUUUAAUAUAGCUAUUGCUAUUGCCAUUGCUGGCAAAUG----------GCUUUAAACACCAGCACUCGAUGGAAACAUGG-CCGAGUUCAGACA (((((....)))))((.(((((...........)))))...((((((((((....)))).))))----------))...........))((((((((....)))..-.)))))....... ( -33.80) >DroEre_CAF1 19710 97 + 1 AGGCCUAAUGGCCUGCACGGCACAACUUUAAUAUAGC------------CAUUGCUGGCAAAUG----------GCUUUAAGCAGCUCCACUCGAUAGAAACACGC-CCGACUUGGGUCA (((((....)))))....(((..........((.(((------------((((.......))))----------))).))...........((....)).....))-).(((....))). ( -24.90) >DroYak_CAF1 19560 108 + 1 AGGCCUAAUGGCCUGCAAGGUACAACUUUAAUGCAGC------------CAUUGCUGGCAAAUGGGCAAAGUGGGCUUUAAGCAGCACCACUCGAUAGAAAUAUUCCCCCAGUUCGGUCA .((((.((((((.(((((((.....)))...))))))------------))))(((((.....(((...((((((((......))).))))).((((....))))))))))))..)))). ( -36.70) >consensus AGGCCUAAUGGCCUGCAAGCUACAACUUUAAUAUAGCUAUUGCUAUUGCCAUUGCUGGCAAAUG__________GCUUUAAGCACCAGCACUCGAUAGAAACAUGG_CCGAGUUCAGACA (((((....)))))....(((............((((....((....))....)))).................((.....))...)))(((((..............)))))....... (-14.84 = -15.40 + 0.56)


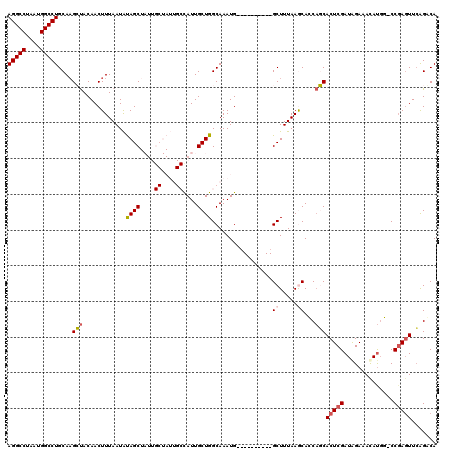
| Location | 5,575,756 – 5,575,865 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -35.18 |
| Consensus MFE | -17.58 |
| Energy contribution | -19.46 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5575756 109 - 22407834 UGCCUGAACUCGG-CCAUGUUUCUAUCGAGUGCUGGUGCUUAAAGC----------CAUUUGCCAGCAAUAGCAAUAGCAAUAGCUAUAUUAAAGUUGUAGCUUGCAGGCCAUUAGGCCU .((((((....((-((.((((.((((....(((((((((.....))----------.....))))))))))).))))(((..(((((((.......)))))))))).)))).)))))).. ( -38.50) >DroSec_CAF1 19799 109 - 1 UGUCUGAACUCGG-CCAUGUUUCCAUCGAGUGCUGGUGUUUAAAGC----------CAUUUGCCAGCAAUGGCAAUAGCAAUAGCUAUAUUAAAGUUGUAGCUUGCAGGCCAUUAGGCCU .((((((....((-((.((((.((((....((((((((........----------....)))))))))))).))))(((..(((((((.......)))))))))).)))).)))))).. ( -36.90) >DroSim_CAF1 21451 109 - 1 UGUCUGAACUCGG-CCAUGUUUCCAUCGAGUGCUGGUGUUUAAAGC----------CAUUUGCCAGCAAUGGCAAUAGCAAUAGCUAUAUUAAAGUUGUAGCUUGCAGGCCAUUAGGCCU .((((((....((-((.((((.((((....((((((((........----------....)))))))))))).))))(((..(((((((.......)))))))))).)))).)))))).. ( -36.90) >DroEre_CAF1 19710 97 - 1 UGACCCAAGUCGG-GCGUGUUUCUAUCGAGUGGAGCUGCUUAAAGC----------CAUUUGCCAGCAAUG------------GCUAUAUUAAAGUUGUGCCGUGCAGGCCAUUAGGCCU .(((....))).(-(((.(((((........))))).((.....))----------....)))).(((.((------------((.(((.......))))))))))(((((....))))) ( -27.80) >DroYak_CAF1 19560 108 - 1 UGACCGAACUGGGGGAAUAUUUCUAUCGAGUGGUGCUGCUUAAAGCCCACUUUGCCCAUUUGCCAGCAAUG------------GCUGCAUUAAAGUUGUACCUUGCAGGCCAUUAGGCCU ....((((.(((((((.....)))...((((((.(((......)))))))))..))))))))...((((.(------------(.((((.......))))))))))(((((....))))) ( -35.80) >consensus UGACUGAACUCGG_CCAUGUUUCUAUCGAGUGCUGGUGCUUAAAGC__________CAUUUGCCAGCAAUGGCAAUAGCAAUAGCUAUAUUAAAGUUGUAGCUUGCAGGCCAUUAGGCCU .......((((((............)))))).....(((....................((((((....))))))........((((((.......))))))..)))((((....)))). (-17.58 = -19.46 + 1.88)

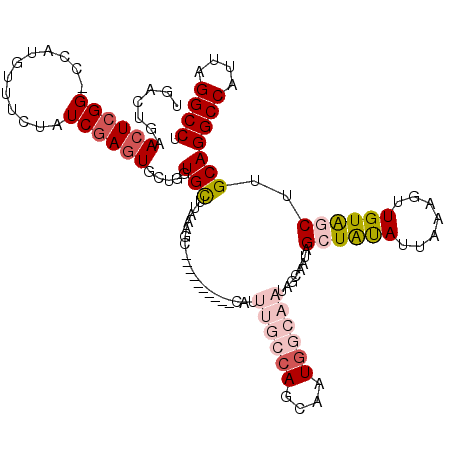
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:42 2006