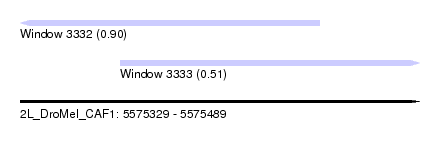
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,575,329 – 5,575,489 |
| Length | 160 |
| Max. P | 0.898043 |

| Location | 5,575,329 – 5,575,449 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -27.80 |
| Energy contribution | -27.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
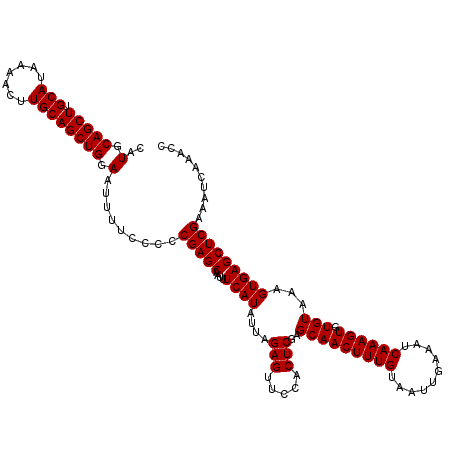
>2L_DroMel_CAF1 5575329 120 - 22407834 CAUGCAGCUUGCAUAAAACUUGCAGCUGGAAUUUUCCCCCGAGCAAUUUCAUAUUAGAGUUCCACUCGAGCAACUUUGUAAUUGAAAUCAAAGUGUGUAAAGUGAGCUCGAAAUCAAACC ..(.(((((.(((.......)))))))).).........(((((....((((....(((.....)))..(((((((((..........)))))).)))...))))))))).......... ( -27.80) >DroSec_CAF1 19381 120 - 1 CAUGCAGCUUGCAUAAAACUUGCAGCUGGAAUUUUCCCCCGAGCAAUUUCAUAUUAGAGUUCCACUCGAGCAACUUUGUAAUUGAAAUCAAAGUGUGUAAAGUGAGCUCGAAAUCAAGCC ...((((((.(((.......)))))))((((((((.....(((....))).....))))))))..((((((.((((((((.(((....)))....))))))))..))))))......)). ( -28.80) >DroSim_CAF1 21033 120 - 1 CAUGCAGCUUGCAUAAAACUUGCAGCUGGAAUUUUCCCCCGAGCAAUUUCAUAUUAGAGUUCCACUCGAGCAACUUUGUAAUUGAAAUCAAAGUGUGUAAAGUGAGCUCGAAAUCAAACC ..(.(((((.(((.......)))))))).).........(((((....((((....(((.....)))..(((((((((..........)))))).)))...))))))))).......... ( -27.80) >DroEre_CAF1 19310 120 - 1 CAUGCAGCUUGCAUAAAACUUGCAGCUGGAAUUUUCCCCCGAGCAAUUUCAUAUUAGAGUUCCACUCGAGCAACUUUGUAAUUGAAAUCAAAGUGUGUAAAGUGAGCUCGAAAUCAAACC ..(.(((((.(((.......)))))))).).........(((((....((((....(((.....)))..(((((((((..........)))))).)))...))))))))).......... ( -27.80) >DroYak_CAF1 19150 120 - 1 CAUGCAGCUUGCAUAAAACUUGCAGCUGGAAUUUUCCCCCGAGCAAUUUCAUAUUAGAGUUCCACUCGAGCAACUUUGUAAUUGAAAUCAAAGUGUGUAAAGUGAGCUCGAAAUCAAACC ..(.(((((.(((.......)))))))).).........(((((....((((....(((.....)))..(((((((((..........)))))).)))...))))))))).......... ( -27.80) >consensus CAUGCAGCUUGCAUAAAACUUGCAGCUGGAAUUUUCCCCCGAGCAAUUUCAUAUUAGAGUUCCACUCGAGCAACUUUGUAAUUGAAAUCAAAGUGUGUAAAGUGAGCUCGAAAUCAAACC ..(.(((((.(((.......)))))))).).........(((((....((((....(((.....)))..(((((((((..........)))))).)))...))))))))).......... (-27.80 = -27.80 + 0.00)

| Location | 5,575,369 – 5,575,489 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.92 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -24.88 |
| Energy contribution | -25.48 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
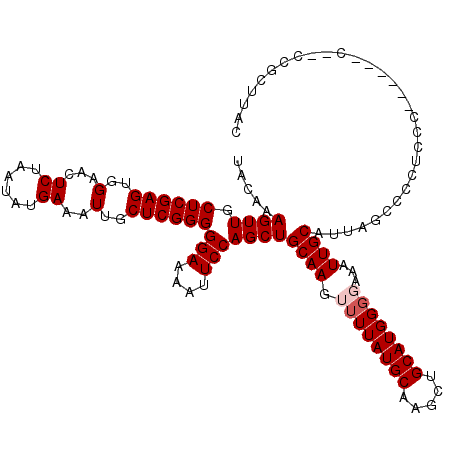
>2L_DroMel_CAF1 5575369 120 + 22407834 UACAAAGUUGCUCGAGUGGAACUCUAAUAUGAAAUUGCUCGGGGGAAAAUUCCAGCUGCAAGUUUUAUGCAAGCUGCAUGGGGAAAUUGCAUUAGCCCCUCCCACCGCCCAGCCGCUUAC ....((((.(((.(((..(...((......))..)..)))((((((......((((((((.......))).)))))...((((..(......)..)))))))).))....))).)))).. ( -35.70) >DroSec_CAF1 19421 114 + 1 UACAAAGUUGCUCGAGUGGAACUCUAAUAUGAAAUUGCUCGGGGGAAAAUUCCAGCUGCAAGUUUUAUGCAAGCUGCAUGGGCAAAUUGCAUUAGCCCCUCCC------CCUCCGCUUAC .............(((((((..((......))........((((((......((((((((.......))).)))))...((((...........)))).))))------))))))))).. ( -35.90) >DroSim_CAF1 21073 114 + 1 UACAAAGUUGCUCGAGUGGAACUCUAAUAUGAAAUUGCUCGGGGGAAAAUUCCAGCUGCAAGUUUUAUGCAAGCUGCAUGGGCAAAUUGCAUUAGCCCCUCCC------CCGCCGCUUAC ....((((.((..(((..(...((......))..)..)))((((((......((((((((.......))).)))))...((((...........)))).))))------)))).)))).. ( -38.60) >DroEre_CAF1 19350 107 + 1 UACAAAGUUGCUCGAGUGGAACUCUAAUAUGAAAUUGCUCGGGGGAAAAUUCCAGCUGCAAGUUUUAUGCAAGCUGCAUGGGGAAAUUGCAUUUGCCCCUC-------------GGUGGC ......((..((((((..(...((......))..)..))))((((.......(((.(((((.((((((((.....))))))))...))))).)))))))..-------------))..)) ( -30.91) >DroYak_CAF1 19190 107 + 1 UACAAAGUUGCUCGAGUGGAACUCUAAUAUGAAAUUGCUCGGGGGAAAAUUCCAGCUGCAAGUUUUAUGCAAGCUGCAUGGCGAAAUUGCAUUAACCCCUC-------------GUUGAC ..(((.......((((..(...((......))..)..))))((((.......((((((((.......))).)))))....(((....))).....))))..-------------.))).. ( -25.90) >consensus UACAAAGUUGCUCGAGUGGAACUCUAAUAUGAAAUUGCUCGGGGGAAAAUUCCAGCUGCAAGUUUUAUGCAAGCUGCAUGGGGAAAUUGCAUUAGCCCCUCCC______C__CCGCUUAC .....((((.((((((..(...((......))..)..))))))(((....)))))))((((.((((((((.....))))))))...)))).............................. (-24.88 = -25.48 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:36 2006