
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,574,703 – 5,574,900 |
| Length | 197 |
| Max. P | 0.927119 |

| Location | 5,574,703 – 5,574,820 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 98.86 |
| Mean single sequence MFE | -31.11 |
| Consensus MFE | -31.26 |
| Energy contribution | -31.04 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5574703 117 + 22407834 GUGGCAGUUGCUGCAGCUGUGUGCGUGUGCUGGGGAUUUGGCAAAUUGAUUUGAGAUUUACUAUUAAGAUGAUUUCCAUGCAUUUUGCAUGCUGUUGCCCAAAUUAUUUUCAUAUUA ..(((....)))(((((.(((((((.(((((((..((((((.(((((.......))))).))).......)))..))).))))..))))))).)))))................... ( -30.91) >DroSec_CAF1 18781 117 + 1 GCGGCAGUUGCUGCAGCUGUGUGCGUGUGCUGGGGAUUUAGCAAAUUGAUUUGAGAUUUACUAUUAAGAUGAUUUCCAUGCAUUUUGCAUGCUGUUGCCCAAAUUAUUUUCAUAUUA (((((....)))))(((.(((((((.(((((((..((((((.(((((.......))))).))).......)))..))).))))..))))))).)))..................... ( -31.51) >DroSim_CAF1 20424 117 + 1 GUGGCAGUUGCUGCAGCUGUGUGCGUGUGCUGGGGAUUUGGCAAAUUGAUUUGAGAUUUACUAUUAAGAUGAUUUCCAUGCAUUUUGCAUGCUGUUGCCCAAAUUAUUUUCAUAUUA ..(((....)))(((((.(((((((.(((((((..((((((.(((((.......))))).))).......)))..))).))))..))))))).)))))................... ( -30.91) >consensus GUGGCAGUUGCUGCAGCUGUGUGCGUGUGCUGGGGAUUUGGCAAAUUGAUUUGAGAUUUACUAUUAAGAUGAUUUCCAUGCAUUUUGCAUGCUGUUGCCCAAAUUAUUUUCAUAUUA ..(((....)))(((((.(((((((.(((((((..((((((.(((((.......))))).))).......)))..))).))))..))))))).)))))................... (-31.26 = -31.04 + -0.22)



| Location | 5,574,740 – 5,574,860 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -20.46 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.60 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5574740 120 - 22407834 GGGCAUUAAUUAGUUUCGUAUUCAUUACAAAAUCUAUAAUUAAUAUGAAAAUAAUUUGGGCAACAGCAUGCAAAAUGCAUGGAAAUCAUCUUAAUAGUAAAUCUCAAAUCAAUUUGCCAA .((((........(((((((((.((((.........)))).)))))))))...(((((((......(((((.....)))))((.....))............))))))).....)))).. ( -21.40) >DroSec_CAF1 18818 120 - 1 GGGCAUUAAUUAGUUUCGUAUUCAUUACAAAAUCUAUAAUUAAUAUGAAAAUAAUUUGGGCAACAGCAUGCAAAAUGCAUGGAAAUCAUCUUAAUAGUAAAUCUCAAAUCAAUUUGCUAA ..((...(((((.(((((((((.((((.........)))).))))))))).)))))...)).....(((((.....))))).............((((((((.........)))))))). ( -22.90) >DroSim_CAF1 20461 120 - 1 GGGCAUUAAUUAGUUUCGUAUUCAUUACAAAAUCUAUAAUUAAUAUGAAAAUAAUUUGGGCAACAGCAUGCAAAAUGCAUGGAAAUCAUCUUAAUAGUAAAUCUCAAAUCAAUUUGCCAA .((((........(((((((((.((((.........)))).)))))))))...(((((((......(((((.....)))))((.....))............))))))).....)))).. ( -21.40) >DroEre_CAF1 18730 119 - 1 -GUCAUUAAUUAGUUUCGUAUUCAUUACAAAAUCUAUAAUUAAUAUGAAAAUAAUUUGGGCAACAGCAUGCAAAAUGCAUGGAAAUCAUCUUAAUAGUAAAUCUCAAAUCAAUUUGCCAA -(((...(((((.(((((((((.((((.........)))).))))))))).)))))..))).....(((((.....)))))...............((((((.........))))))... ( -20.20) >DroYak_CAF1 18585 119 - 1 -GGCAUUAAUUAGUUUCGUAUUCAUUACAAAAUCUAUAAUUAAUAUGAAAAUAAUUUGGGCAACAGCAUGCAAAAUUCAUGGAAAUCAUCUUAAUAGUAAAUCUCAAAUCAAUUUGCCAA -((((........(((((((((.((((.........)))).)))))))))...(((((((......((((.......))))((.....))............))))))).....)))).. ( -16.40) >consensus GGGCAUUAAUUAGUUUCGUAUUCAUUACAAAAUCUAUAAUUAAUAUGAAAAUAAUUUGGGCAACAGCAUGCAAAAUGCAUGGAAAUCAUCUUAAUAGUAAAUCUCAAAUCAAUUUGCCAA .......(((((.(((((((((.((((.........)))).))))))))).)))))..(((((...(((((.....)))))((.((...((....))...)).))........))))).. (-18.56 = -18.60 + 0.04)

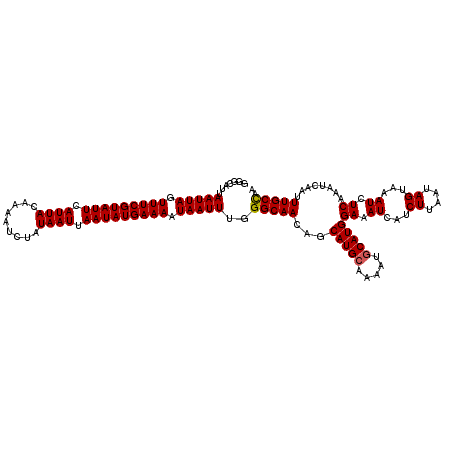
| Location | 5,574,780 – 5,574,900 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.66 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -21.40 |
| Energy contribution | -22.44 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5574780 120 - 22407834 GCUUUCGCAAAAUGCCAUGCCUUCUUGUCAUUGAAUUUAGGGGCAUUAAUUAGUUUCGUAUUCAUUACAAAAUCUAUAAUUAAUAUGAAAAUAAUUUGGGCAACAGCAUGCAAAAUGCAU ((....))....(((((((((((...((......))...))))))).(((((.(((((((((.((((.........)))).))))))))).)))))..))))...((((.....)))).. ( -25.50) >DroSec_CAF1 18858 120 - 1 GCUUGAGCCAAAUGCCAUGCCUUCUUGUCAUUGAAUUCGGGGGCAUUAAUUAGUUUCGUAUUCAUUACAAAAUCUAUAAUUAAUAUGAAAAUAAUUUGGGCAACAGCAUGCAAAAUGCAU (((...(((.((((((.((..(((........)))..))..))))))(((((.(((((((((.((((.........)))).))))))))).)))))..)))...)))((((.....)))) ( -28.40) >DroSim_CAF1 20501 120 - 1 GCUUGCGCCAAAUGCCAUGCCUUCUUGUCAUUGAAUUCGGGGGCAUUAAUUAGUUUCGUAUUCAUUACAAAAUCUAUAAUUAAUAUGAAAAUAAUUUGGGCAACAGCAUGCAAAAUGCAU ((((((.((((((...((((((((..((......))..)))))))).......(((((((((.((((.........)))).)))))))))...)))))))))..)))((((.....)))) ( -30.20) >DroEre_CAF1 18770 119 - 1 GCAUGCGCCAAAUGCCAUGCCUUCUUGUCAUUGAAUUUGG-GUCAUUAAUUAGUUUCGUAUUCAUUACAAAAUCUAUAAUUAAUAUGAAAAUAAUUUGGGCAACAGCAUGCAAAAUGCAU ((((((.((((((.(.(((.(.....).))).).))))))-(((...(((((.(((((((((.((((.........)))).))))))))).)))))..)))....))))))......... ( -30.50) >DroYak_CAF1 18625 119 - 1 CCAAUCGCAAUAUGUCAUGCCUUCUUGUCAUUGAAUUUAG-GGCAUUAAUUAGUUUCGUAUUCAUUACAAAAUCUAUAAUUAAUAUGAAAAUAAUUUGGGCAACAGCAUGCAAAAUUCAU ......(((...(((..(((((...((((...........-))))..(((((.(((((((((.((((.........)))).))))))))).))))).))))))))...)))......... ( -20.90) >consensus GCUUGCGCCAAAUGCCAUGCCUUCUUGUCAUUGAAUUUGGGGGCAUUAAUUAGUUUCGUAUUCAUUACAAAAUCUAUAAUUAAUAUGAAAAUAAUUUGGGCAACAGCAUGCAAAAUGCAU ............((((((((((((...((...))....)))))))).(((((.(((((((((.((((.........)))).))))))))).)))))..))))...((((.....)))).. (-21.40 = -22.44 + 1.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:34 2006