
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,574,431 – 5,574,625 |
| Length | 194 |
| Max. P | 0.995642 |

| Location | 5,574,431 – 5,574,545 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.82 |
| Mean single sequence MFE | -53.07 |
| Consensus MFE | -38.30 |
| Energy contribution | -41.18 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.95 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5574431 114 + 22407834 GGCCGUCAGGGGCAUGAUAAAUGUCCUCAAUUGCAGAUCCUGCCUAUCUGCGGCAUGAAUCUCCUACGCAGUUGCAGU------UGCAAUUGCAACUGCAGUUGCAUGCAGGAUGGCCUG (((((((.(((((((.....)))))))...(((((((((.((((.......)))).).)))......(((..((((((------(((....)))))))))..))).)))))))))))).. ( -55.40) >DroSec_CAF1 18516 108 + 1 GGCCGUCAGGGGCAUGAUAAAUGUCCUCAAUUGCAGAUCCUGCCUAUCUGCGGCAUGAAUCUCCUACGCAGUUGCAAC------UG------CAACUGCAGUUGCAUGCAGGAUGGCCUG ((((....(((((((.....))))))).........(((((((....(((((..............))))).((((((------((------(....))))))))).))))))))))).. ( -48.84) >DroSim_CAF1 20166 114 + 1 GGCCGUCAGGGGCAUGAUAAAUGUCCUCAAUUGCAGAUCCUGCCUAUCUGCGGCAUGAAUCUCCUACGCAGUUGCAAC------UGCAACUGCAACUGCAGUUGCAUGCAGGAUGGCCUG (((((((.(((((((.....)))))))(((((((((.((.((((.......)))).)).........((((((((...------.))))))))..))))))))).......))))))).. ( -54.60) >DroEre_CAF1 18433 120 + 1 GCCCGUCAGGGGCAUGAUAAAUGUCCUCAAUUGCAGAUCCUGCCUAUCUGCGGGAUGCAUCUCCUACGCAGUUGCAGCUGCAGUUGCAGUUGCAGCUACAGUUGCAUGCAGGAUGGCCUG (((.....(((((((.....))))))).........(((((((....((((((((......)))..))))).((((((((.((((((....)))))).)))))))).))))))))))... ( -52.70) >DroYak_CAF1 18276 111 + 1 GGCCGUCAGGGGCAUGAUAAAUGUCCUCAAUUGCAGAUCCUGCCUAUCUGCAGGAUGAAUCUCCUACGA---------CGCAGUUGCAGCUGCAGCUGCAGUUGCAUGCAGGAUGGCCUG (((((((.(((((((.....)))))))...(((((.(((((((......)))))))..........(((---------(((((((((....)))))))).))))..)))))))))))).. ( -53.80) >consensus GGCCGUCAGGGGCAUGAUAAAUGUCCUCAAUUGCAGAUCCUGCCUAUCUGCGGCAUGAAUCUCCUACGCAGUUGCAAC______UGCAACUGCAACUGCAGUUGCAUGCAGGAUGGCCUG (((((((.(((((((.....)))))))(((((((((...((((......))))..............(((((((((........)))))))))..))))))))).......))))))).. (-38.30 = -41.18 + 2.88)


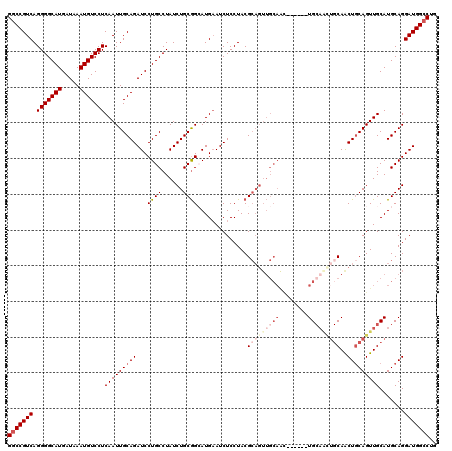
| Location | 5,574,431 – 5,574,545 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.82 |
| Mean single sequence MFE | -45.68 |
| Consensus MFE | -28.16 |
| Energy contribution | -29.56 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5574431 114 - 22407834 CAGGCCAUCCUGCAUGCAACUGCAGUUGCAAUUGCA------ACUGCAACUGCGUAGGAGAUUCAUGCCGCAGAUAGGCAGGAUCUGCAAUUGAGGACAUUUAUCAUGCCCCUGACGGCC ..((((.((((((..(((..(((((((((....)))------))))))..)))))))))(((((.((((.......)))))))))........(((.(((.....)))..)))...)))) ( -46.90) >DroSec_CAF1 18516 108 - 1 CAGGCCAUCCUGCAUGCAACUGCAGUUG------CA------GUUGCAACUGCGUAGGAGAUUCAUGCCGCAGAUAGGCAGGAUCUGCAAUUGAGGACAUUUAUCAUGCCCCUGACGGCC ..((((.((((((((((((((((....)------))------))))))..)))...(.((((((.((((.......)))))))))).).....))))......(((......))).)))) ( -44.00) >DroSim_CAF1 20166 114 - 1 CAGGCCAUCCUGCAUGCAACUGCAGUUGCAGUUGCA------GUUGCAACUGCGUAGGAGAUUCAUGCCGCAGAUAGGCAGGAUCUGCAAUUGAGGACAUUUAUCAUGCCCCUGACGGCC ..((((.((((((.(((....)))...((((((((.------...))))))))))))))(((((.((((.......)))))))))........(((.(((.....)))..)))...)))) ( -46.00) >DroEre_CAF1 18433 120 - 1 CAGGCCAUCCUGCAUGCAACUGUAGCUGCAACUGCAACUGCAGCUGCAACUGCGUAGGAGAUGCAUCCCGCAGAUAGGCAGGAUCUGCAAUUGAGGACAUUUAUCAUGCCCCUGACGGGC ...((.(((((((.((((.((((((.(((....))).)))))).)))).(((((..(((......))))))))....)))))))..))...................((((.....)))) ( -46.60) >DroYak_CAF1 18276 111 - 1 CAGGCCAUCCUGCAUGCAACUGCAGCUGCAGCUGCAACUGCG---------UCGUAGGAGAUUCAUCCUGCAGAUAGGCAGGAUCUGCAAUUGAGGACAUUUAUCAUGCCCCUGACGGCC ..((((((((((((((((..((((((....))))))..))))---------).((((((......))))))......))))))).........(((.(((.....)))..)))...)))) ( -44.90) >consensus CAGGCCAUCCUGCAUGCAACUGCAGUUGCAACUGCA______GCUGCAACUGCGUAGGAGAUUCAUGCCGCAGAUAGGCAGGAUCUGCAAUUGAGGACAUUUAUCAUGCCCCUGACGGCC ..((((.((((((.((((((....))))))...............((....))))))))..........((((((.......)))))).....(((.(((.....)))..)))...)))) (-28.16 = -29.56 + 1.40)


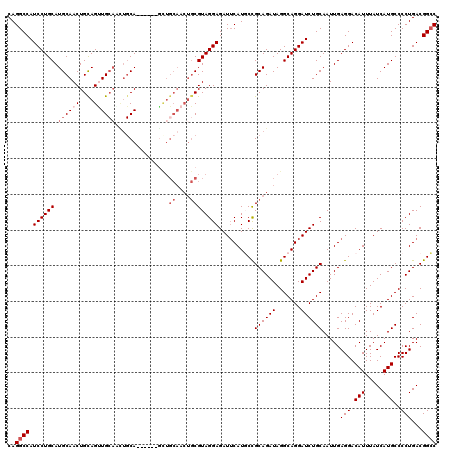
| Location | 5,574,471 – 5,574,585 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.63 |
| Mean single sequence MFE | -43.08 |
| Consensus MFE | -26.09 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5574471 114 - 22407834 AGGCAAAGUCGACGGCAUUUACAUAAACUUCGUGGACACUCAGGCCAUCCUGCAUGCAACUGCAGUUGCAAUUGCA------ACUGCAACUGCGUAGGAGAUUCAUGCCGCAGAUAGGCA .(((...(((.((((..(((....)))..)))).)))......))).((((((..(((..(((((((((....)))------))))))..)))))))))......((((.......)))) ( -43.50) >DroSec_CAF1 18556 107 - 1 G-GCAAAGUCGGCGGCAUUUACAUAAACUUCGUGGACACUCAGGCCAUCCUGCAUGCAACUGCAGUUG------CA------GUUGCAACUGCGUAGGAGAUUCAUGCCGCAGAUAGGCA .-((...(((.(((((((.........((.((..(.....((((....))))..(((((((((....)------))------)))))).)..)).)).......))))))).)))..)). ( -41.49) >DroSim_CAF1 20206 114 - 1 GGGCAAAGUCGACGGCAUUUACAUAAACUUCGUGGACACUCAGGCCAUCCUGCAUGCAACUGCAGUUGCAGUUGCA------GUUGCAACUGCGUAGGAGAUUCAUGCCGCAGAUAGGCA .(((...(((.((((..(((....)))..)))).)))......))).((((((.(((....)))...((((((((.------...))))))))))))))......((((.......)))) ( -42.40) >DroEre_CAF1 18473 120 - 1 AGGCAAAGUCGACGGCAUUUACAUAAACUUCGUGGACACUCAGGCCAUCCUGCAUGCAACUGUAGCUGCAACUGCAACUGCAGCUGCAACUGCGUAGGAGAUGCAUCCCGCAGAUAGGCA .......(((.((((..(((....)))..)))).)))......((((((.((((((((..((((((((((........))))))))))..))))).(((......))).)))))).))). ( -43.90) >DroYak_CAF1 18316 111 - 1 GGCCAAAGUCGACGGCAUUUACAUAAACUUCGUGGACACUCAGGCCAUCCUGCAUGCAACUGCAGCUGCAGCUGCAACUGCG---------UCGUAGGAGAUUCAUCCUGCAGAUAGGCA .(((...(((.((((..(((....)))..)))).))).((((((...(((((((((((..((((((....))))))..))))---------).)))))).......)))).))...))). ( -44.10) >consensus GGGCAAAGUCGACGGCAUUUACAUAAACUUCGUGGACACUCAGGCCAUCCUGCAUGCAACUGCAGUUGCAACUGCA______GCUGCAACUGCGUAGGAGAUUCAUGCCGCAGAUAGGCA .......(((.((((..(((....)))..)))).)))......((((((.((((((((..(((((((((....)))......))))))..))))).((.........)))))))).))). (-26.09 = -26.90 + 0.81)

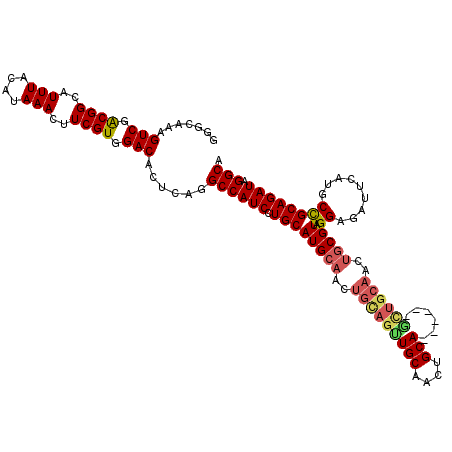
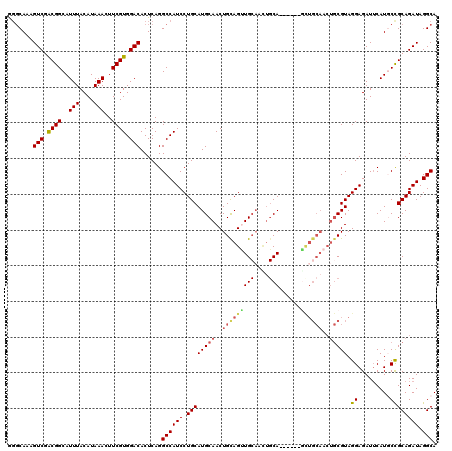
| Location | 5,574,509 – 5,574,625 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.42 |
| Mean single sequence MFE | -37.68 |
| Consensus MFE | -31.82 |
| Energy contribution | -32.30 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5574509 116 - 22407834 CUAAAGUAGGCAGCAAGCAGGAAUCAGGACGACGUCAUCGAGGCAAAGUCGACGGCAUUUACAUAAACUUCGUGGACACUCAGGCCAUCCUGCAUGCAACUGCAGUUGCAAUUGCA---- ............((((((((((.......(((.....))).(((...(((.((((..(((....)))..)))).)))......))).)))))).((((((....)))))).)))).---- ( -35.80) >DroSec_CAF1 18594 109 - 1 CUAAAGUAGGCAGCAAGCAGGAAUCAGGACGACGUCAUCGG-GCAAAGUCGGCGGCAUUUACAUAAACUUCGUGGACACUCAGGCCAUCCUGCAUGCAACUGCAGUUG------CA---- .........(((((..((((((.......(((.....)))(-((...(((.((((..(((....)))..)))).)))......))).)))))).(((....)))))))------).---- ( -34.30) >DroSim_CAF1 20244 116 - 1 CUAAAGUAGGCAGCAAGCAGGAAUCAGGACGACGUCAUCGGGGCAAAGUCGACGGCAUUUACAUAAACUUCGUGGACACUCAGGCCAUCCUGCAUGCAACUGCAGUUGCAGUUGCA---- .........(((((..((((((.......(((.....))).(((...(((.((((..(((....)))..)))).)))......))).)))))).((((((....))))))))))).---- ( -39.10) >DroEre_CAF1 18513 120 - 1 CUAAAGUAGGCAGCAGGCAGGAAUCAGGACGACGUCAUCGAGGCAAAGUCGACGGCAUUUACAUAAACUUCGUGGACACUCAGGCCAUCCUGCAUGCAACUGUAGCUGCAACUGCAACUG .....(((((((((..((((((.......(((.....))).(((...(((.((((..(((....)))..)))).)))......))).)))))).(((....))))))))..))))..... ( -37.20) >DroYak_CAF1 18347 120 - 1 CUAAAGUAGGCAGCAGGCAGGAAUCAGGACGACGUCAUCGGGCCAAAGUCGACGGCAUUUACAUAAACUUCGUGGACACUCAGGCCAUCCUGCAUGCAACUGCAGCUGCAGCUGCAACUG .....(((((((((..((((((.......(((.....)))((((...(((.((((..(((....)))..)))).))).....)))).)))))).(((....))))))))..))))..... ( -42.00) >consensus CUAAAGUAGGCAGCAAGCAGGAAUCAGGACGACGUCAUCGGGGCAAAGUCGACGGCAUUUACAUAAACUUCGUGGACACUCAGGCCAUCCUGCAUGCAACUGCAGUUGCAACUGCA____ .....(((((((((..((((((.......(((.....))).(((...(((.((((..(((....)))..)))).)))......))).)))))).(((....))))))))..))))..... (-31.82 = -32.30 + 0.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:31 2006