
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,572,697 – 5,572,849 |
| Length | 152 |
| Max. P | 0.994293 |

| Location | 5,572,697 – 5,572,809 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.03 |
| Mean single sequence MFE | -33.31 |
| Consensus MFE | -26.04 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
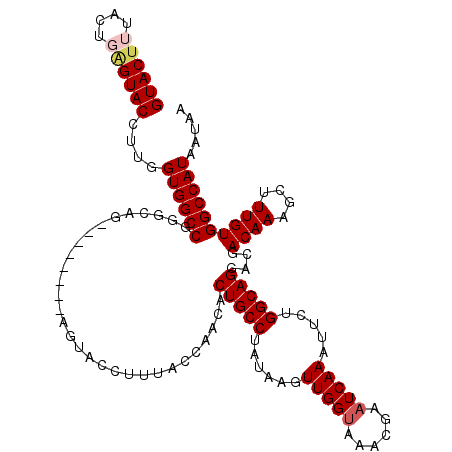
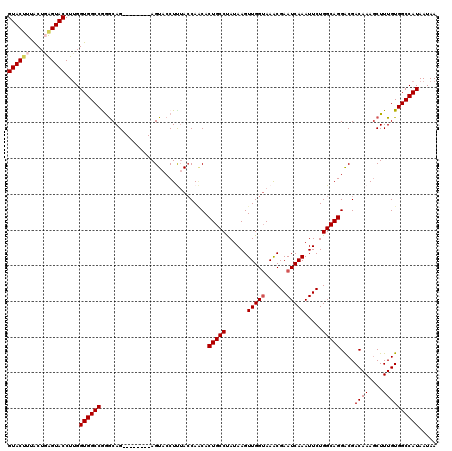
>2L_DroMel_CAF1 5572697 112 - 22407834 GUACUUUACUGAGUACCUUGGUGGCCAGACAG--------AGUACCUUUACCAACACUGCCUAUAAGUUGGUAAACGAAUCAAAUUCUGGCAGGGCGACAAACCUUUGUGGCCAUAAUAA ((((((....)))))).......((((((..(--------(...(.(((((((((...........))))))))).)..))....))))))..(((.((((....)))).)))....... ( -33.80) >DroSec_CAF1 16728 112 - 1 GUACUUUACUGAGUACCUUGGUGGCCGGGCAG--------AGUACCUUUACCAACACUGCCUAUAAGUUGGUAAACGAAUCAAAUUCUGGCAGGACGACAAAGCUUUGUGGCCAUAAUAA ((((((....))))))....((((((..((((--------((...((((.......(((((......(((((......))))).....)))))......))))))))))))))))..... ( -32.22) >DroSim_CAF1 18103 112 - 1 GUACUUUACUGAGUACCUUGGUGGCCGGGCAG--------AGUACCUUUACCAACACUGCCUAUAAGUUGGUAAACGAAUCAAAUUCUGGCAGGACGACAAAGCUUUGUGGCCAUAAUAA ((((((....))))))....((((((..((((--------((...((((.......(((((......(((((......))))).....)))))......))))))))))))))))..... ( -32.22) >DroEre_CAF1 16731 120 - 1 GUACCUUGCUCAGUACCUUGGUGGCCAGGCAGCGGCAAAUAGUACCUCUACCAUCACUGCCCAUAAGUUGGGAAACGAAUCAAAUUCUGGCAGGACGACAAAGCUUUGUGGCCAUAAUAA ((((........))))....(((((((.(.(((((....(((.....))))).((.(((((.....(((.(....).)))........))))))).......))).).)))))))..... ( -31.32) >DroYak_CAF1 16566 120 - 1 GUACUUUACUCGGUACCUGGGUGGCCGAGCAGAGGCGAAUAGUACCUCAAACAGCACUGCCUAUAAGUUGGUAAGCGAAUCAAAUUCUGGCAGGGCGACAAAGCUUUGUGGCCAUAAUAA (((((......)))))....((((((((((.((((.........)))).....((.(((((......(((((......))))).....))))).))......))))...))))))..... ( -37.00) >consensus GUACUUUACUGAGUACCUUGGUGGCCGGGCAG________AGUACCUUUACCAACACUGCCUAUAAGUUGGUAAACGAAUCAAAUUCUGGCAGGACGACAAAGCUUUGUGGCCAUAAUAA ((((((....))))))....((((((..............................(((((......(((((......))))).....)))))....((((....))))))))))..... (-26.04 = -26.72 + 0.68)

| Location | 5,572,737 – 5,572,849 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.88 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -17.78 |
| Energy contribution | -18.90 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.891108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
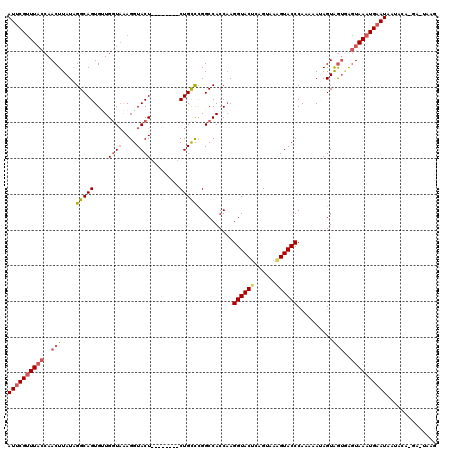
>2L_DroMel_CAF1 5572737 112 + 22407834 AUUCGUUUACCAACUUAUAGGCAGUGUUGGUAAAGGUACU--------CUGUCUGGCCACCAAGGUACUCAGUAAAGUACCCAAAAAUAGUAGUAGGUAAAUGCAUAAUACACGAUUAAG ...((((((((.(((..(((((((.((..........)).--------)))))))........((((((......))))))..........))).))))))))................. ( -25.90) >DroSec_CAF1 16768 101 + 1 AUUCGUUUACCAACUUAUAGGCAGUGUUGGUAAAGGUACU--------CUGCCCGGCCACCAAGGUACUCAGUAAAGUACCCAAAAAUAGUGGUGGGUAAAUGAAUUAU----------- (((((((((((........(((((.((..........)).--------)))))....(((((.((((((......)))))).........))))))))))))))))...----------- ( -33.30) >DroSim_CAF1 18143 101 + 1 AUUCGUUUACCAACUUAUAGGCAGUGUUGGUAAAGGUACU--------CUGCCCGGCCACCAAGGUACUCAGUAAAGUACCCAAAAAUAGUAAAUAGUAAAUGAAUAAU----------- ((((((((((.........(((((.((..........)).--------)))))..........((((((......))))))...............))))))))))...----------- ( -24.70) >DroEre_CAF1 16771 119 + 1 AUUCGUUUCCCAACUUAUGGGCAGUGAUGGUAGAGGUACUAUUUGCCGCUGCCUGGCCACCAAGGUACUGAGCAAGGUACCCAAA-AAAGUGGUGAAUAAAUGAAUAGCACAUGAUUAAG ((((((((..........(((((((...((((((.......))))))))))))).(((((...((((((......))))))....-...)))))....)))))))).............. ( -33.20) >DroYak_CAF1 16606 120 + 1 AUUCGCUUACCAACUUAUAGGCAGUGCUGUUUGAGGUACUAUUCGCCUCUGCUCGGCCACCCAGGUACCGAGUAAAGUACCCAAAAAAAGUAGUGAGUAAAUGAAUAAUACAUGAAUAAG (((..(((((..((((...((..(((((....(((((.......)))))(((((((..((....)).))))))).))))))).....)))).)))))..))).................. ( -29.80) >consensus AUUCGUUUACCAACUUAUAGGCAGUGUUGGUAAAGGUACU________CUGCCCGGCCACCAAGGUACUCAGUAAAGUACCCAAAAAUAGUAGUGAGUAAAUGAAUAAUACA_GA_UAAG ((((((((((..(((....(((((....((((....))))........)))))..........((((((......))))))..........)))..)))))))))).............. (-17.78 = -18.90 + 1.12)


| Location | 5,572,737 – 5,572,849 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.88 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -20.96 |
| Energy contribution | -21.32 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
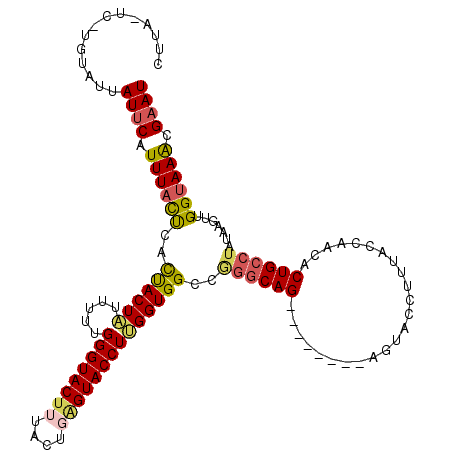
>2L_DroMel_CAF1 5572737 112 - 22407834 CUUAAUCGUGUAUUAUGCAUUUACCUACUACUAUUUUUGGGUACUUUACUGAGUACCUUGGUGGCCAGACAG--------AGUACCUUUACCAACACUGCCUAUAAGUUGGUAAACGAAU .....(((((((((.((..(((..((((((........((((((((....))))))))))))))..))))).--------)))))..((((((((...........)))))))))))).. ( -27.60) >DroSec_CAF1 16768 101 - 1 -----------AUAAUUCAUUUACCCACCACUAUUUUUGGGUACUUUACUGAGUACCUUGGUGGCCGGGCAG--------AGUACCUUUACCAACACUGCCUAUAAGUUGGUAAACGAAU -----------...((((.((((((..((((((.....((((((((....))))))))))))))..((((((--------.((..........)).)))))).......)))))).)))) ( -33.50) >DroSim_CAF1 18143 101 - 1 -----------AUUAUUCAUUUACUAUUUACUAUUUUUGGGUACUUUACUGAGUACCUUGGUGGCCGGGCAG--------AGUACCUUUACCAACACUGCCUAUAAGUUGGUAAACGAAU -----------...((((.((((((((((((((((...((((((((....)))))))).)))))..((((((--------.((..........)).)))))).)))).))))))).)))) ( -29.70) >DroEre_CAF1 16771 119 - 1 CUUAAUCAUGUGCUAUUCAUUUAUUCACCACUUU-UUUGGGUACCUUGCUCAGUACCUUGGUGGCCAGGCAGCGGCAAAUAGUACCUCUACCAUCACUGCCCAUAAGUUGGGAAACGAAU .........((((((((........(((((....-...((((((........)))))))))))(((.......))).))))))))........((....((((.....))))....)).. ( -29.21) >DroYak_CAF1 16606 120 - 1 CUUAUUCAUGUAUUAUUCAUUUACUCACUACUUUUUUUGGGUACUUUACUCGGUACCUGGGUGGCCGAGCAGAGGCGAAUAGUACCUCAAACAGCACUGCCUAUAAGUUGGUAAGCGAAU ...((((..(((((((((...((((((..........))))))((((.((((((((....)).)))))).))))..)))))))))........((..((((........)))).)))))) ( -33.60) >consensus CUUA_UC_UGUAUUAUUCAUUUACUCACUACUAUUUUUGGGUACUUUACUGAGUACCUUGGUGGCCGGGCAG________AGUACCUUUACCAACACUGCCUAUAAGUUGGUAAACGAAU ..............((((.((((((..((((((.....((((((((....))))))))))))))..((((((........................)))))).......)))))).)))) (-20.96 = -21.32 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:25 2006