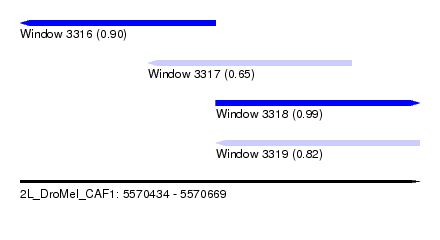
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,570,434 – 5,570,669 |
| Length | 235 |
| Max. P | 0.991348 |

| Location | 5,570,434 – 5,570,549 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.64 |
| Mean single sequence MFE | -38.04 |
| Consensus MFE | -30.38 |
| Energy contribution | -29.78 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5570434 115 - 22407834 UAACUUCCCCACUCGGGUCGCAGCAAAUUAUGAAGAUGGCCAUCGCAUCCUGGCGAUUAGUAAUUUGAUCAG-UAUCUAUUGAUUGAUGGCCAGGUGGCCG----UCCGGCUGAGUCAUU ..........((((((.(((((........))..((((((((......((((((.((((((.....(((...-.))).....)))))).))))))))))))----))))))))))).... ( -38.20) >DroSec_CAF1 15271 116 - 1 UAACUUCCCCACUCGGGUCGCAGCAAAUUAUGAAGAUGGCCAUCGCAUCCUGGCGAUUAGUAAUUUGAUUAUCUAUCUAUUGAUUGAUGGCCAGGUGGCCG----UCCGGCUGAGUCAUU ..........((((((.(((((........))..((((((((......((((((.((((((.....(((.....))).....)))))).))))))))))))----))))))))))).... ( -39.00) >DroSim_CAF1 16833 116 - 1 UAACUUCCCCACUCGGGUCGCAGCAAAUUAUGAAGAUGGCCAUCGCAUCCUGGCGAUUAGUAAUUUGAUCAUCUAUCUAUUGAUUGAUGGCCAGGUGGCCG----UCCGGCUGAGUCAUU ..........((((((.(((((........))..((((((((......((((((((((((....))))))(((.(((....))).))).))))))))))))----))))))))))).... ( -39.10) >DroEre_CAF1 15386 117 - 1 UAACUCGCCCACUUGGGUCGCAGCAAAUUAUGAAGAUGGCCAUCGCAUCCUGGCGAUUAGUAAUUUGAUCAG-UAUCUAUUGAUUGAUGACCAGGUGGCUG--AGUCCGGCGGAGUCAUU ..(((((.((((((((.(((((.((((((((...(((.((((........)))).))).))))))))(((((-(....)))))))).))))))))))).))--)))..(((...)))... ( -38.10) >DroYak_CAF1 15526 119 - 1 UAACUUCCCCACUUGGGUCGCAGCAAAUUAUGAAGAUGGCCAUCGCAUCCUGGCGAUUACUAAUUUGAUCAG-UAUCUAUUGAUUGAUGGCCAGGUGGUCGUCCGUUCGGCUGAGUCAUU .......(((....)))...((((.....(((..((((((((......((((((.(((........((((((-(....)))))))))).)))))))))))))))))...))))....... ( -35.80) >consensus UAACUUCCCCACUCGGGUCGCAGCAAAUUAUGAAGAUGGCCAUCGCAUCCUGGCGAUUAGUAAUUUGAUCAG_UAUCUAUUGAUUGAUGGCCAGGUGGCCG____UCCGGCUGAGUCAUU ..((((...(((...((((....((((((((...(((.((((........)))).))).))))))))((((...(((....)))))))))))..)))((((......)))).)))).... (-30.38 = -29.78 + -0.60)

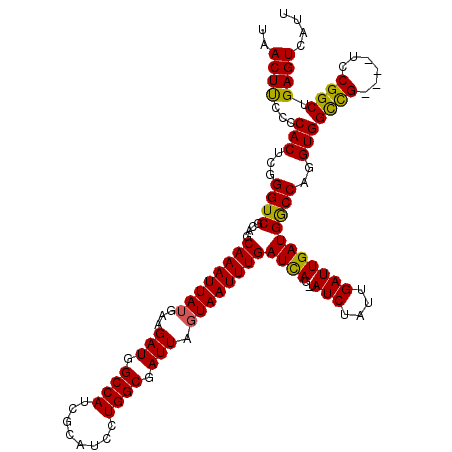
| Location | 5,570,509 – 5,570,629 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -24.50 |
| Energy contribution | -25.10 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5570509 120 - 22407834 CAUGAAUGGCAGCUCACAAAUUUGCUGCCUUAAAAUAUUGUCUAAGUGAAAAACUUUUCGCAUAUGAUUUAUUUACUUGAUAACUUCCCCACUCGGGUCGCAGCAAAUUAUGAAGAUGGC ........((..((((..((((((((((....(((((..(((((.((((((....)))))).)).))).))))).............(((....)))..)))))))))).)).))...)) ( -26.40) >DroSec_CAF1 15347 120 - 1 CAUAAAUGGCAGCGCACAAAUUUGCUGCCUUAAAAUAUUGUCUAAGUGAAAAACUUUUCGCAUAUGAUUUAUUUACUUGAUAACUUCCCCACUCGGGUCGCAGCAAAUUAUGAAGAUGGC .......(.((...((..((((((((((....(((((..(((((.((((((....)))))).)).))).))))).............(((....)))..)))))))))).))....)).) ( -25.40) >DroSim_CAF1 16909 120 - 1 CAUGAAUGGCAGCGCACAAAUUUGCUGCCUUAAAAUAUUGUCUAAGUGAAAAACUUUUCGCAUAUGAUUUAUUUACUUGAUAACUUCCCCACUCGGGUCGCAGCAAAUUAUGAAGAUGGC .......(.((...((..((((((((((....(((((..(((((.((((((....)))))).)).))).))))).............(((....)))..)))))))))).))....)).) ( -25.40) >DroEre_CAF1 15463 120 - 1 CAUGAAUGGCAGCUCACAAAUUUGCUGCCUUAAAAUAUUGUCUAAGUGAAAAACUUUUCGCAUAUGAUUUAUUUACUUGAUAACUCGCCCACUUGGGUCGCAGCAAAUUAUGAAGAUGGC ........((..((((..((((((((((....(((((..(((((.((((((....)))))).)).))).)))))............((((....)))).)))))))))).)).))...)) ( -28.20) >DroYak_CAF1 15605 120 - 1 CAUGAAUGGCAGCUCACAAAUUUGCUGCCUUAAAAUAUUGUCUAAGUGAAAAACUUUUCGCAUAUGAUUUAUAUACUCGAUAACUUCCCCACUUGGGUCGCAGCAAAUUAUGAAGAUGGC ........((..((((..((((((((((......((((.(((((.((((((....)))))).)).)))..)))).............(((....)))..)))))))))).)).))...)) ( -26.00) >consensus CAUGAAUGGCAGCUCACAAAUUUGCUGCCUUAAAAUAUUGUCUAAGUGAAAAACUUUUCGCAUAUGAUUUAUUUACUUGAUAACUUCCCCACUCGGGUCGCAGCAAAUUAUGAAGAUGGC .......(.((..(((..((((((((((....(((((..(((((.((((((....)))))).)).))).))))).............(((....)))..)))))))))).)))...)).) (-24.50 = -25.10 + 0.60)

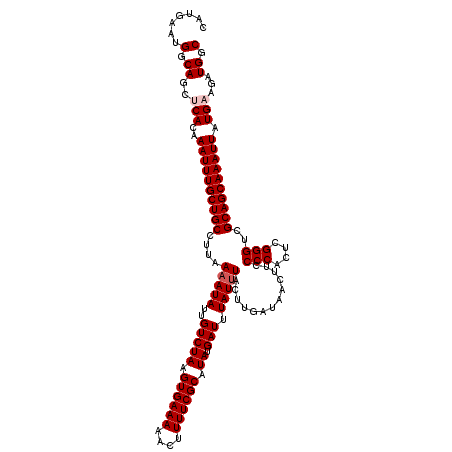
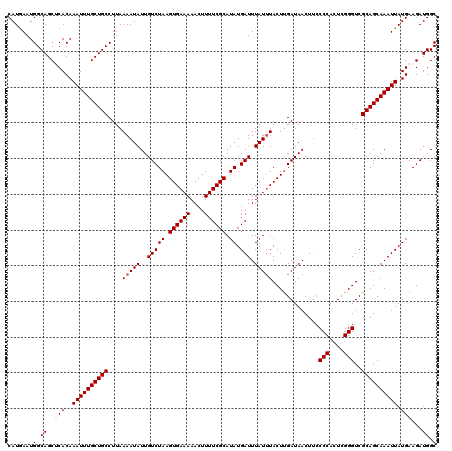
| Location | 5,570,549 – 5,570,669 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -24.78 |
| Energy contribution | -24.62 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5570549 120 + 22407834 UCAAGUAAAUAAAUCAUAUGCGAAAAGUUUUUCACUUAGACAAUAUUUUAAGGCAGCAAAUUUGUGAGCUGCCAUUCAUGCAAGUAUUAUGAUAGCCAUAAGUCUGAGCCAACUUCCCAA ..((((...............((((....)))).(((((((..........((((((..........)))))).............(((((.....))))))))))))...))))..... ( -24.30) >DroSec_CAF1 15387 120 + 1 UCAAGUAAAUAAAUCAUAUGCGAAAAGUUUUUCACUUAGACAAUAUUUUAAGGCAGCAAAUUUGUGCGCUGCCAUUUAUGCAAGUAUUAUGAUGGCCAUAAGUCUGAGCCAACUUCCCAA ..((((......(((((((((((((....))))..................((((((..........))))))..........))).))))))(((((......)).))).))))..... ( -24.60) >DroSim_CAF1 16949 120 + 1 UCAAGUAAAUAAAUCAUAUGCGAAAAGUUUUUCACUUAGACAAUAUUUUAAGGCAGCAAAUUUGUGCGCUGCCAUUCAUGCAAGUAUUAUGAUGGCCAUAAGUCUGAGCCAACUUCCCAA ..((((......(((((((((((((....))))..................((((((..........))))))..........))).))))))(((((......)).))).))))..... ( -24.60) >DroEre_CAF1 15503 120 + 1 UCAAGUAAAUAAAUCAUAUGCGAAAAGUUUUUCACUUAGACAAUAUUUUAAGGCAGCAAAUUUGUGAGCUGCCAUUCAUGCAAGUAUUAUGAUAGCCAUAAGUCUGAGCCAACUUGCCAA .(((((...............((((....)))).(((((((..........((((((..........)))))).............(((((.....))))))))))))...))))).... ( -27.00) >DroYak_CAF1 15645 120 + 1 UCGAGUAUAUAAAUCAUAUGCGAAAAGUUUUUCACUUAGACAAUAUUUUAAGGCAGCAAAUUUGUGAGCUGCCAUUCAUGCUAGUAUUAUGAUAGCCAUAAGUCUGAGCCAACUUGCCAA ....((((((.....))))))(..(((((.....(((((((..........((((((..........))))))......((((.........)))).....)))))))..)))))..).. ( -28.40) >consensus UCAAGUAAAUAAAUCAUAUGCGAAAAGUUUUUCACUUAGACAAUAUUUUAAGGCAGCAAAUUUGUGAGCUGCCAUUCAUGCAAGUAUUAUGAUAGCCAUAAGUCUGAGCCAACUUCCCAA ..((((...............((((....)))).(((((((..........((((((..........)))))).............(((((.....))))))))))))...))))..... (-24.78 = -24.62 + -0.16)

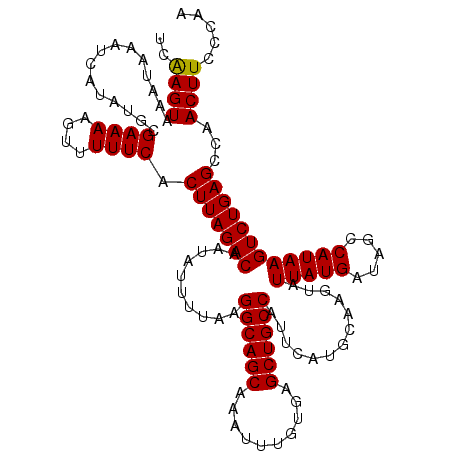
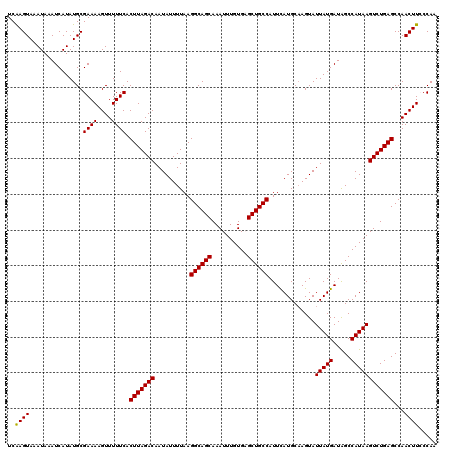
| Location | 5,570,549 – 5,570,669 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -24.42 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.822239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5570549 120 - 22407834 UUGGGAAGUUGGCUCAGACUUAUGGCUAUCAUAAUACUUGCAUGAAUGGCAGCUCACAAAUUUGCUGCCUUAAAAUAUUGUCUAAGUGAAAAACUUUUCGCAUAUGAUUUAUUUACUUGA ..(..(((((.(((.(((((((((.....))))).............((((((..........))))))..........)))).)))....)))))..)..................... ( -26.10) >DroSec_CAF1 15387 120 - 1 UUGGGAAGUUGGCUCAGACUUAUGGCCAUCAUAAUACUUGCAUAAAUGGCAGCGCACAAAUUUGCUGCCUUAAAAUAUUGUCUAAGUGAAAAACUUUUCGCAUAUGAUUUAUUUACUUGA (..((....((((.((......))))))(((((..............(((((((........)))))))................((((((....)))))).)))))........))..) ( -27.40) >DroSim_CAF1 16949 120 - 1 UUGGGAAGUUGGCUCAGACUUAUGGCCAUCAUAAUACUUGCAUGAAUGGCAGCGCACAAAUUUGCUGCCUUAAAAUAUUGUCUAAGUGAAAAACUUUUCGCAUAUGAUUUAUUUACUUGA (..((....((((.((......))))))(((((..............(((((((........)))))))................((((((....)))))).)))))........))..) ( -27.40) >DroEre_CAF1 15503 120 - 1 UUGGCAAGUUGGCUCAGACUUAUGGCUAUCAUAAUACUUGCAUGAAUGGCAGCUCACAAAUUUGCUGCCUUAAAAUAUUGUCUAAGUGAAAAACUUUUCGCAUAUGAUUUAUUUACUUGA ...((((((((((.((......)))))).......))))))......((((((..........))))))...(((((..(((((.((((((....)))))).)).))).)))))...... ( -28.71) >DroYak_CAF1 15645 120 - 1 UUGGCAAGUUGGCUCAGACUUAUGGCUAUCAUAAUACUAGCAUGAAUGGCAGCUCACAAAUUUGCUGCCUUAAAAUAUUGUCUAAGUGAAAAACUUUUCGCAUAUGAUUUAUAUACUCGA ..(((((....(((.((..(((((.....)))))..)))))......((((((..........))))))........)))))...((((((....))))))................... ( -25.30) >consensus UUGGGAAGUUGGCUCAGACUUAUGGCUAUCAUAAUACUUGCAUGAAUGGCAGCUCACAAAUUUGCUGCCUUAAAAUAUUGUCUAAGUGAAAAACUUUUCGCAUAUGAUUUAUUUACUUGA ..((((((((.(((.(((((((((.....))))).............((((((..........))))))..........)))).)))....))))))))..................... (-24.42 = -24.82 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:22 2006