
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,553,662 – 5,553,851 |
| Length | 189 |
| Max. P | 0.999071 |

| Location | 5,553,662 – 5,553,777 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.98 |
| Mean single sequence MFE | -39.68 |
| Consensus MFE | -33.20 |
| Energy contribution | -33.32 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5553662 115 + 22407834 UAAUUGUCAAACUCCAAUACGCUAAAGUCCACGCUCUUUCCCUGGGUGGCCAUCUU-----AGUCCUACUGCUGUGUGGUGUGGAUGGACUUCAGCCAAGAGCAGGUUCCUGCUCCUCUU ....................(((.((((((((((((.......)))))(((((..(-----(((......)))).))))).....))))))).)))...((((((....))))))..... ( -36.70) >DroSec_CAF1 5826 115 + 1 UAAUUGUCGAACUCCAAUACGCUGAAGUCCACGCUCUUUCCCUGGGUGGCCAUACU-----AAUCCUACUGCUGUGUGGUGUGGAUGGACUUCAGCCAAGAGCAGAUUCCUGCUCCUUUG ....................((((((((((((((((.......))))).(((((((-----(....(((....))))))))))).)))))))))))...((((((....))))))..... ( -43.60) >DroSim_CAF1 5943 115 + 1 UAAUUGUCAAACUCCAAUACGCUGAAGUCCACGCUCUUUCCCUGGGUGGCCAUACU-----AAUCCUACUGCUGUGUGGUGUGGAUGGACUUCAGCCAAGAGCAGGUUCCUGCUCCUUGG .............((((...((((((((((((((((.......))))).(((((((-----(....(((....))))))))))).)))))))))))...((((((....)))))).)))) ( -47.00) >DroEre_CAF1 6016 115 + 1 UAGUUGUCAAACUCCAAUACGCUGAAGUCCACGUUCUUUCCCUGUGAGCCCAUUCUGCUGCAAUCCUACUGCU----GGUGUGGAUGGACUUCAGCCAAGAGCAGGUCCUUGCUC-UUUG .((((....)))).......(((((((((((.((((.........))))((((.((...(((.......))).----)).)))).))))))))))).((((((((....))))))-)).. ( -38.40) >DroYak_CAF1 6155 110 + 1 UAGUUGUCAAACUCCAAUACGCUGAAGUCCACAUUCUUUCCCUGUGAGCCCAUCCU-----AAUCCUACUGCU----GGUGUAGAUGGACUUCAGCCAAGAGCAGGUCCUUGCUC-UUUG .((((....)))).......((((((((((((((.........)))..........-----.......((((.----...)))).))))))))))).((((((((....))))))-)).. ( -32.70) >consensus UAAUUGUCAAACUCCAAUACGCUGAAGUCCACGCUCUUUCCCUGGGUGGCCAUACU_____AAUCCUACUGCUGUGUGGUGUGGAUGGACUUCAGCCAAGAGCAGGUUCCUGCUCCUUUG ..((((........))))..((((((((((((((((.......))))).................((((.(((....))))))).)))))))))))...((((((....))))))..... (-33.20 = -33.32 + 0.12)

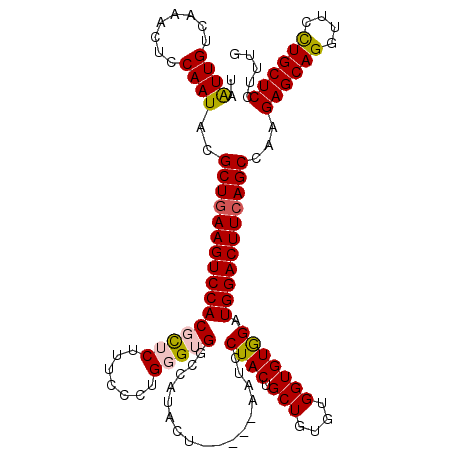
| Location | 5,553,662 – 5,553,777 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.98 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -29.86 |
| Energy contribution | -30.34 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5553662 115 - 22407834 AAGAGGAGCAGGAACCUGCUCUUGGCUGAAGUCCAUCCACACCACACAGCAGUAGGACU-----AAGAUGGCCACCCAGGGAAAGAGCGUGGACUUUAGCGUAUUGGAGUUUGACAAUUA ...((((((((....)))))))).((((((((((((.(...((((......)).)).((-----....(((....))).)).....).))))))))))))..((((........)))).. ( -36.70) >DroSec_CAF1 5826 115 - 1 CAAAGGAGCAGGAAUCUGCUCUUGGCUGAAGUCCAUCCACACCACACAGCAGUAGGAUU-----AGUAUGGCCACCCAGGGAAAGAGCGUGGACUUCAGCGUAUUGGAGUUCGACAAUUA ...((((((((....)))))))).((((((((((((.(...((((......)).))...-----....(((....)))........).))))))))))))..((((........)))).. ( -36.50) >DroSim_CAF1 5943 115 - 1 CCAAGGAGCAGGAACCUGCUCUUGGCUGAAGUCCAUCCACACCACACAGCAGUAGGAUU-----AGUAUGGCCACCCAGGGAAAGAGCGUGGACUUCAGCGUAUUGGAGUUUGACAAUUA (((((((((((....)))))))).((((((((((((.(...((((......)).))...-----....(((....)))........).))))))))))))....)))............. ( -39.90) >DroEre_CAF1 6016 115 - 1 CAAA-GAGCAAGGACCUGCUCUUGGCUGAAGUCCAUCCACACC----AGCAGUAGGAUUGCAGCAGAAUGGGCUCACAGGGAAAGAACGUGGACUUCAGCGUAUUGGAGUUUGACAACUA ..((-(((((......))))))).((((((((((((.....((----.(((((...)))))(((.......)))....))........)))))))))))).......((((....)))). ( -35.82) >DroYak_CAF1 6155 110 - 1 CAAA-GAGCAAGGACCUGCUCUUGGCUGAAGUCCAUCUACACC----AGCAGUAGGAUU-----AGGAUGGGCUCACAGGGAAAGAAUGUGGACUUCAGCGUAUUGGAGUUUGACAACUA ..((-(((((......))))))).((((((((((((((((...----....))))....-----....((......))..........)))))))))))).......((((....)))). ( -32.50) >consensus CAAAGGAGCAGGAACCUGCUCUUGGCUGAAGUCCAUCCACACCACACAGCAGUAGGAUU_____AGGAUGGCCACCCAGGGAAAGAGCGUGGACUUCAGCGUAUUGGAGUUUGACAAUUA ...((((((((....)))))))).((((((((((((((((...........)).)))...........((......))...........)))))))))))...(((........)))... (-29.86 = -30.34 + 0.48)

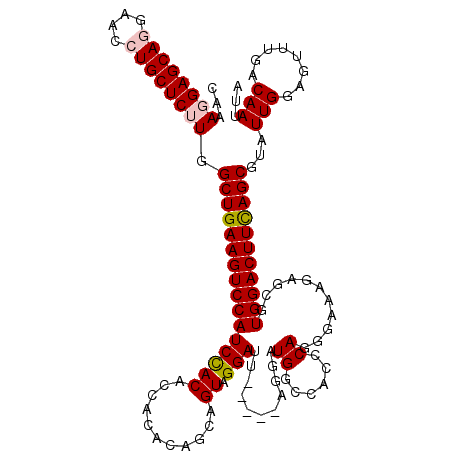

| Location | 5,553,737 – 5,553,851 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.02 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -18.52 |
| Energy contribution | -18.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5553737 114 - 22407834 UAACACACAACACAAACACACACUCUUUUAA------AUAUAACACUUACACAUUAAAAGGGACUUUUUCCUGAACCACAAAGAGGAGCAGGAACCUGCUCUUGGCUGAAGUCCAUCCAC ........................(((((((------................)))))))(((((((..((((.....))...((((((((....))))))))))..)))))))...... ( -25.09) >DroSec_CAF1 5901 109 - 1 -A--ACACGACACAAACA--CACUCUUUUAA------AUAUAACACUCACACAUUAGAAGGGGCUUUUUUCUGAACCGCACAAAGGAGCAGGAAUCUGCUCUUGGCUGAAGUCCAUCCAC -.--....(((.......--..(((((((((------................)))))))))...............((....((((((((....)))))))).))....)))....... ( -22.29) >DroSim_CAF1 6018 112 - 1 UAACACACGACACAAACA--CACUCUUUUUA------AUAUAACACUUACACAUUAGAAAGGACUUUUUCCUGAACCACACCAAGGAGCAGGAACCUGCUCUUGGCUGAAGUCCAUCCAC ..................--.....((((((------((.............))))))))(((((((..((((........))((((((((....))))))))))..)))))))...... ( -24.82) >DroEre_CAF1 6092 106 - 1 UGGCACACGGC--AAAUA--CACUU-UUUAA------ACACAACACUCACACUC--GCAGGGGCUUUUUCCUGAACCACACAAA-GAGCAAGGACCUGCUCUUGGCUGAAGUCCAUCCAC (((.((.((((--.....--.....-.....------.................--.((((((....)))))).........((-(((((......))))))).))))..)))))..... ( -22.00) >DroYak_CAF1 6226 112 - 1 UAGCACACGAC--AAAUA--CACUU-CUUUAUGAAUUAAAAAACACUCACACAU--AAAGGGGCUUUUUCCUGAACCACACAAA-GAGCAAGGACCUGCUCUUGGCUGAAGUCCAUCUAC ...........--.....--.....-(((((((..................)))--))))(((((((..((((.....))..((-(((((......)))))))))..)))))))...... ( -22.17) >consensus UAACACACGACACAAACA__CACUCUUUUAA______AUAUAACACUCACACAUUAGAAGGGGCUUUUUCCUGAACCACACAAAGGAGCAGGAACCUGCUCUUGGCUGAAGUCCAUCCAC ............................................................(((((((..((((.....))...((((((((....))))))))))..)))))))...... (-18.52 = -18.92 + 0.40)

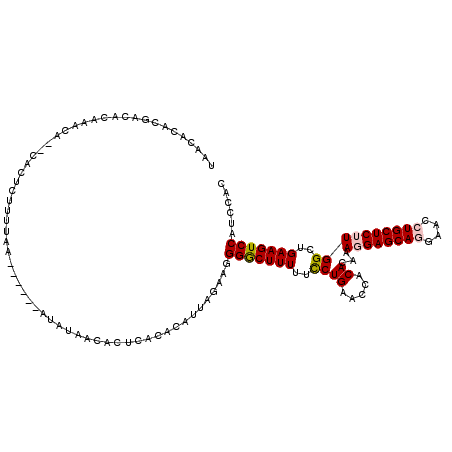
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:09 2006