| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,536,527 – 5,536,639 |
| Length | 112 |
| Max. P | 0.920236 |

| Location | 5,536,527 – 5,536,639 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -30.55 |
| Consensus MFE | -21.31 |
| Energy contribution | -21.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5536527 112 - 22407834 UGCAAGCGGUCGCAGUGCAUCAAGAACUACUGCGACUGCUAUCAGUCCAUGGCCAUCUGCACCAAGUUCUGUCGAUGCGUUGGUAUGAUUUCAUUUCUAUAAGCAUAAAUAU (((.(((((((((((((..........)))))))))))))((((..(((..((((((.(((........))).)))).)))))..)))).............)))....... ( -34.90) >DroSec_CAF1 10566 108 - 1 UGCAAGCGGUCGCAGUGCAUCAAGAACUACUGCGACUGCUACCAGUCCAUGGCCAUCUGCACCAAGUUCUGUCGAUGCACUGGUAGGAUUUCAUUUCUAUAAACA----UAU ....(((((((((((((..........)))))))))))))((((((.((((((...((......))....))).))).))))))((((......)))).......----... ( -34.80) >DroEre_CAF1 10589 104 - 1 UGCAAGAGGUCGCAGUGCAUCAAGAACUACUGCGAUUGCUACCAGUCCAUGGCGAUCUGCACCAAGUUCUGUCGAUGCAUCGGUAAGAUUUCACUUCUA----CA----UAC .....((((((((.(((((((.((((((..(((((((((((........)))))))).)))...))))))...)))))))..))..)))))).......----..----... ( -33.50) >DroYak_CAF1 10574 104 - 1 UGCAAGAGGUCGCAGUGCAUCAAGAACUACUGCGAUUGCUAUCAAUCCAUGGCGAUCUGCACCAUGUUCUGUCGCUGCAUUGGUAAGAUUUCUCUUCUA----CA----UAU ....(((((((.(((((((.(.(((((...((((((((((((......))))))))).)))....)))))...).)))))))....)))))))......----..----... ( -32.90) >DroMoj_CAF1 10556 97 - 1 UGCAAGCGCUCGCAAUGCAUCAAGAACUACUGCGAUUGCUAUCAGUCCAUGGCCAUUUGCUCCAUCUACUGCAAAUGCGUUGGUAAGCUC-------AA----UU----GGC .((.((((.(((((................))))).))))......(((..(((((((((..........))))))).)))))...))..-------..----..----... ( -21.69) >DroAna_CAF1 10307 105 - 1 UGCAAGCGGUCUCAGUGCAUCAAAAACUACUGCGACUGCUAUCAAUCCAUGGUGAUUUGCUCCAAGUUCUGCCGCUGUGUCGGUAAGUAUAUGUUUCAAA---AG----GAA ....(((((((.(((((..........))))).))))))).....(((.(((.(.....).))).....(((((......)))))...............---.)----)). ( -25.50) >consensus UGCAAGCGGUCGCAGUGCAUCAAGAACUACUGCGACUGCUAUCAGUCCAUGGCCAUCUGCACCAAGUUCUGUCGAUGCAUUGGUAAGAUUUCAUUUCUA____CA____UAU .....((((((((((((..........))))))))))))(((((((.((((((.....((.....))...)))).)).)))))))........................... (-21.31 = -21.32 + 0.00)

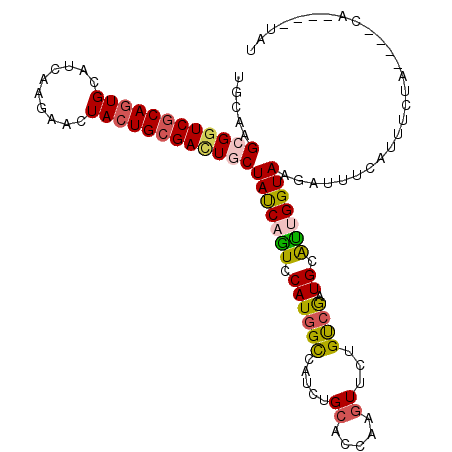

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:00 2006