
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,515,536 – 5,515,708 |
| Length | 172 |
| Max. P | 0.845499 |

| Location | 5,515,536 – 5,515,654 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 84.01 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5515536 118 - 22407834 CUAAUUAGGACUAGUAACAAAAGGUGCUGGCAGUCUGGAAAAAUUGUAAUUCAGUUGAUAAGAAGUAAUUGGCACUCAGCACCUUACUUUAUCUAUUAUAUAUUCUGUUUAUAUUCCA .......(((.((((((...((((((((((((((.(....).)))))......((..((........))..))...)))))))))...))).)))..(((((.......)))))))). ( -24.90) >DroSim_CAF1 146004 108 - 1 CUAAUUAGGACUAGCACCAAAAGGUGCUGGCAGUCUGGAAAAAUUGUAAUUCAGCUGAUAAGAAGUAAGUGGCACUCAGCACCUUACCUUAUCUAG----------GCUGAUAUUCCA .......(((((((((((....))))))))((((.(....).))))....((((((((((((..(((((..((.....))..)))))))))))..)----------)))))...))). ( -36.00) >DroYak_CAF1 144760 105 - 1 CUAAUUAGGUUGUG---CAAAAGGUGCUGGGAGGCUGGAAAAAUUGUAAUUCAGUUGAUAAGAAGUAAGUGCCACUCAGCACCUUACAUUAUCUAU----------GCUGAUAUUCCA .....(((((.(((---...(((((((((((.(((......(((((.....)))))..............))).))))))))))).))).))))).----------............ ( -28.85) >consensus CUAAUUAGGACUAG_A_CAAAAGGUGCUGGCAGUCUGGAAAAAUUGUAAUUCAGUUGAUAAGAAGUAAGUGGCACUCAGCACCUUACAUUAUCUAU__________GCUGAUAUUCCA .......(((..........((((((((((..(((........((((..........)))).........)))..))))))))))....((((................)))).))). (-18.40 = -18.62 + 0.22)



| Location | 5,515,614 – 5,515,708 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.10 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -13.39 |
| Energy contribution | -14.06 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5515614 94 + 22407834 UUCCAGACUGCCAGCACCUUUUGUUACUAGUCCUAAUUAGCAGGACACUGCCAUGGAG---CCAUAUUCCCGCCUCCUGGCUGCUCCUUGCUU---------------CAAA .....(((((..((((.....))))..)))))......(((((((....((((.((((---.(........).))))))))...))).)))).---------------.... ( -25.10) >DroSim_CAF1 146072 94 + 1 UUCCAGACUGCCAGCACCUUUUGGUGCUAGUCCUAAUUAGCAGGACACUGCCAUGGAG---CCGUAUACCCACCUACUGGCUGCUCCUUGCUC---------------CAAA .....((((...((((((....)))))))))).......((((....))))..(((((---(........((((....)).))......))))---------------)).. ( -28.64) >DroYak_CAF1 144828 109 + 1 UUCCAGCCUCCCAGCACCUUUUG---CACAACCUAAUUAGCAGGACACUGCCAUGGAAUUCCCGAAUUCCCGCAUCCUGGCUCCUCCUUGCUCCUGUAUCCUCGCAGUCCAA ....((((.....(((.....))---)..............((((...(((...((((((....)))))).)))))))))))...........((((......))))..... ( -20.90) >consensus UUCCAGACUGCCAGCACCUUUUG_U_CUAGUCCUAAUUAGCAGGACACUGCCAUGGAG___CCGUAUUCCCGCCUCCUGGCUGCUCCUUGCUC_______________CAAA ......................................(((((((....((((.((((...............))))))))...))).)))).................... (-13.39 = -14.06 + 0.67)


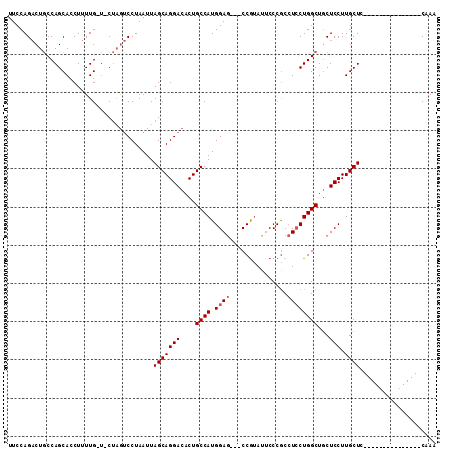
| Location | 5,515,614 – 5,515,708 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.10 |
| Mean single sequence MFE | -32.51 |
| Consensus MFE | -18.38 |
| Energy contribution | -19.50 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
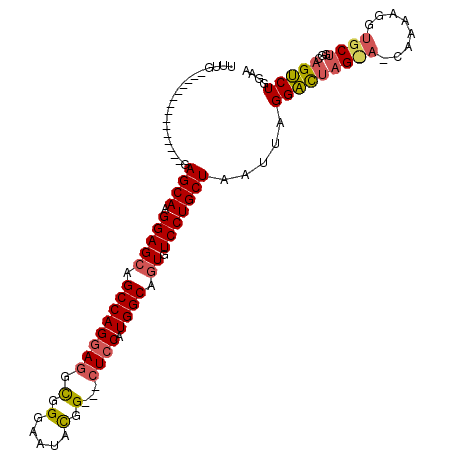
>2L_DroMel_CAF1 5515614 94 - 22407834 UUUG---------------AAGCAAGGAGCAGCCAGGAGGCGGGAAUAUGG---CUCCAUGGCAGUGUCCUGCUAAUUAGGACUAGUAACAAAAGGUGCUGGCAGUCUGGAA ....---------------.((((.(((((.((((((((.((......)).---)))).)))).)).)))))))..(((((.((((((.(....).))))))...))))).. ( -31.10) >DroSim_CAF1 146072 94 - 1 UUUG---------------GAGCAAGGAGCAGCCAGUAGGUGGGUAUACGG---CUCCAUGGCAGUGUCCUGCUAAUUAGGACUAGCACCAAAAGGUGCUGGCAGUCUGGAA ..((---------------((((.....((.(((....)))..)).....)---)))))((((((....)))))).(((((.((((((((....))))))))...))))).. ( -36.70) >DroYak_CAF1 144828 109 - 1 UUGGACUGCGAGGAUACAGGAGCAAGGAGGAGCCAGGAUGCGGGAAUUCGGGAAUUCCAUGGCAGUGUCCUGCUAAUUAGGUUGUG---CAAAAGGUGCUGGGAGGCUGGAA (..(.((.(.((.((......((((.((..(((.(((((((.((((((....))))))......))))))))))..))...)))).---......)).)).).)).)..).. ( -29.72) >consensus UUUG_______________GAGCAAGGAGCAGCCAGGAGGCGGGAAUACGG___CUCCAUGGCAGUGUCCUGCUAAUUAGGACUAG_A_CAAAAGGUGCUGGCAGUCUGGAA ....................((((.(((((.((((((((.(.(.....).)...)))).)))).)).))))))).....(((((((((........))))...))))).... (-18.38 = -19.50 + 1.12)

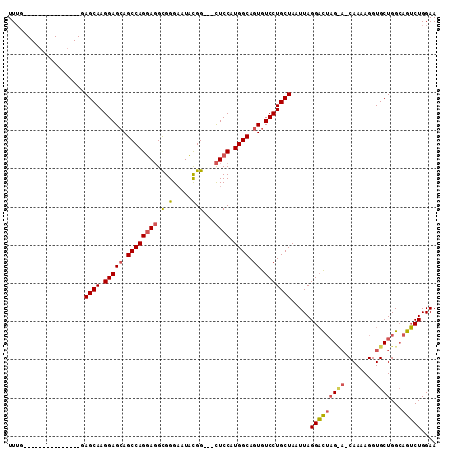
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:52 2006